This function utilizes the Seurat package to perform a differential expression (DE) test on gene expression data. Users have the flexibility to specify custom cell groups, marker types, and various options for DE analysis.
Usage
RunDEtest(
srt,
group.by = NULL,
group1 = NULL,
group2 = NULL,
cells1 = NULL,
cells2 = NULL,
features = NULL,
markers_type = c("all", "paired", "conserved", "disturbed"),
grouping.var = NULL,
meta.method = c("maximump", "minimump", "wilkinsonp", "meanp", "sump", "votep"),
test.use = "wilcox",
only.pos = TRUE,
fc.threshold = 1.5,
base = 2,
pseudocount.use = 1,
mean.fxn = NULL,
min.pct = 0.1,
min.diff.pct = -Inf,
max.cells.per.ident = Inf,
latent.vars = NULL,
min.cells.feature = 3,
min.cells.group = 3,
norm.method = "LogNormalize",
p.adjust.method = "bonferroni",
layer = "data",
assay = NULL,
seed = 11,
verbose = TRUE,
cores = 1,
...
)Arguments
- srt
A Seurat object.
- group.by
A grouping variable in the dataset to define the groups or conditions for the differential test. If not provided, the function uses the "active.ident" variable in the Seurat object.
- group1
A vector of cell IDs or a character vector specifying the cells that belong to the first group. If both group.by and group1 are provided, group1 takes precedence.
- group2
A vector of cell IDs or a character vector specifying the cells that belong to the second group. This parameter is only used when group.by or group1 is provided.
- cells1
A vector of cell IDs specifying the cells that belong to group1. If provided, group1 is ignored.
- cells2
A vector of cell IDs specifying the cells that belong to group2. This parameter is only used when cells1 is provided.
- features
A vector of gene names specifying the features to consider for the differential test. If not provided, all features in the dataset are considered.
- markers_type
A character value specifying the type of markers to find. Possible values are "all", "paired", "conserved", and "disturbed".
- grouping.var
A character value specifying the grouping variable for finding conserved or disturbed markers. This parameter is only used when markers_type is "conserved" or "disturbed".
- meta.method
A character value specifying the method to use for combining p-values in the conserved markers test. Possible values are "maximump", "minimump", "wilkinsonp", "meanp", "sump", and "votep".
- test.use
Denotes which test to use. Available options are:
"wilcox" : Identifies differentially expressed genes between two groups of cells using a Wilcoxon Rank Sum test (default); will use a fast implementation by Presto if installed
"wilcox_limma" : Identifies differentially expressed genes between two groups of cells using the limma implementation of the Wilcoxon Rank Sum test; set this option to reproduce results from Seurat v4
"bimod" : Likelihood-ratio test for single cell gene expression, (McDavid et al., Bioinformatics, 2013)
"roc" : Identifies 'markers' of gene expression using ROC analysis. For each gene, evaluates (using AUC) a classifier built on that gene alone, to classify between two groups of cells. An AUC value of 1 means that expression values for this gene alone can perfectly classify the two groupings (i.e. Each of the cells in cells.1 exhibit a higher level than each of the cells in cells.2). An AUC value of 0 also means there is perfect classification, but in the other direction. A value of 0.5 implies that the gene has no predictive power to classify the two groups. Returns a 'predictive power' (abs(AUC-0.5) * 2) ranked matrix of putative differentially expressed genes.
"t" : Identify differentially expressed genes between two groups of cells using the Student's t-test.
"negbinom" : Identifies differentially expressed genes between two groups of cells using a negative binomial generalized linear model. Use only for UMI-based datasets
"poisson" : Identifies differentially expressed genes between two groups of cells using a poisson generalized linear model. Use only for UMI-based datasets
"LR" : Uses a logistic regression framework to determine differentially expressed genes. Constructs a logistic regression model predicting group membership based on each feature individually and compares this to a null model with a likelihood ratio test.
"MAST" : Identifies differentially expressed genes between two groups of cells using a hurdle model tailored to scRNA-seq data. Utilizes the MAST package to run the DE testing.
"DESeq2" : Identifies differentially expressed genes between two groups of cells based on a model using DESeq2 which uses a negative binomial distribution (Love et al, Genome Biology, 2014).This test does not support pre-filtering of genes based on average difference (or percent detection rate) between cell groups. However, genes may be pre-filtered based on their minimum detection rate (min.pct) across both cell groups. To use this method, please install DESeq2, using the instructions at https://bioconductor.org/packages/release/bioc/html/DESeq2.html
- only.pos
Only return positive markers (FALSE by default)
- fc.threshold
A numeric value used to filter genes for testing based on their average fold change between/among the two groups. Default is
1.5.- base
The base with respect to which logarithms are computed.
- pseudocount.use
Pseudocount to add to averaged expression values when calculating logFC. 1 by default.
- mean.fxn
Function to use for fold change or average difference calculation. The default depends on the the value of
fc.slot:"counts" : difference in the log of the mean counts, with pseudocount.
"data" : difference in the log of the average exponentiated data, with pseudocount. This adjusts for differences in sequencing depth between cells, and assumes that "data" has been log-normalized.
"scale.data" : difference in the means of scale.data.
- min.pct
only test genes that are detected in a minimum fraction of min.pct cells in either of the two populations. Meant to speed up the function by not testing genes that are very infrequently expressed. Default is 0.01
- min.diff.pct
only test genes that show a minimum difference in the fraction of detection between the two groups. Set to -Inf by default
- max.cells.per.ident
Down sample each identity class to a max number. Default is no downsampling. Not activated by default (set to Inf)
- latent.vars
Variables to test, used only when
test.useis one of 'LR', 'negbinom', 'poisson', or 'MAST'- min.cells.feature
Minimum number of cells expressing the feature in at least one of the two groups, currently only used for poisson and negative binomial tests
- min.cells.group
Minimum number of cells in one of the groups
- norm.method
Normalization method for fold change calculation when layer is 'data'. Default is
"LogNormalize".- p.adjust.method
A character value specifying the method to use for adjusting p-values. Default is
"bonferroni".- layer
Which layer to use. Default is
data.- assay
Assay to use in differential expression testing
- seed
Random seed for reproducibility. Default is
11.- verbose
Whether to print the message. Default is
TRUE.- cores
The number of cores to use for parallelization with foreach::foreach. Default is
1.- ...
Additional arguments to pass to the Seurat::FindMarkers function.
Examples
data(pancreas_sub)
pancreas_sub <- standard_scop(pancreas_sub)
#> ℹ [2026-02-11 03:49:36] Start standard scop workflow...
#> ℹ [2026-02-11 03:49:36] Checking a list of <Seurat>...
#> ! [2026-02-11 03:49:36] Data 1/1 of the `srt_list` is "unknown"
#> ℹ [2026-02-11 03:49:36] Perform `NormalizeData()` with `normalization.method = 'LogNormalize'` on the data 1/1 of the `srt_list`...
#> ℹ [2026-02-11 03:49:38] Perform `Seurat::FindVariableFeatures()` on the data 1/1 of the `srt_list`...
#> ℹ [2026-02-11 03:49:39] Use the separate HVF from srt_list
#> ℹ [2026-02-11 03:49:39] Number of available HVF: 2000
#> ℹ [2026-02-11 03:49:39] Finished check
#> ℹ [2026-02-11 03:49:39] Perform `Seurat::ScaleData()`
#> ℹ [2026-02-11 03:49:40] Perform pca linear dimension reduction
#> ℹ [2026-02-11 03:49:41] Perform `Seurat::FindClusters()` with `cluster_algorithm = 'louvain'` and `cluster_resolution = 0.6`
#> ℹ [2026-02-11 03:49:41] Reorder clusters...
#> ℹ [2026-02-11 03:49:41] Perform umap nonlinear dimension reduction
#> ℹ [2026-02-11 03:49:41] Non-linear dimensionality reduction (umap) using (Standardpca) dims (1-50) as input
#> ℹ [2026-02-11 03:49:45] Non-linear dimensionality reduction (umap) using (Standardpca) dims (1-50) as input
#> ✔ [2026-02-11 03:49:49] Run scop standard workflow completed
pancreas_sub <- RunDEtest(
pancreas_sub,
group.by = "SubCellType"
)
#> ℹ [2026-02-11 03:49:49] Data type is log-normalized
#> ℹ [2026-02-11 03:49:49] Start differential expression test
#> ℹ [2026-02-11 03:49:49] Find all markers(wilcox) among [1] 8 groups...
#> ℹ [2026-02-11 03:49:49] Using 1 core
#> ⠙ [2026-02-11 03:49:49] Running for Ductal [1/8] ■■■■■ …
#> ✔ [2026-02-11 03:49:49] Completed 8 tasks in 1.2s
#>
#> ℹ [2026-02-11 03:49:49] Building results
#> ✔ [2026-02-11 03:49:50] Differential expression test completed
AllMarkers <- dplyr::filter(
pancreas_sub@tools$DEtest_SubCellType$AllMarkers_wilcox,
p_val_adj < 0.05 & avg_log2FC > 1
)
ht1 <- GroupHeatmap(
pancreas_sub,
features = AllMarkers$gene,
feature_split = AllMarkers$group1,
group.by = "SubCellType"
)
#> ℹ [2026-02-11 03:49:53] The size of the heatmap is fixed because certain elements are not scalable.
#> ℹ [2026-02-11 03:49:53] The width and height of the heatmap are determined by the size of the current viewport.
#> ℹ [2026-02-11 03:49:53] If you want to have more control over the size, you can manually set the parameters 'width' and 'height'.
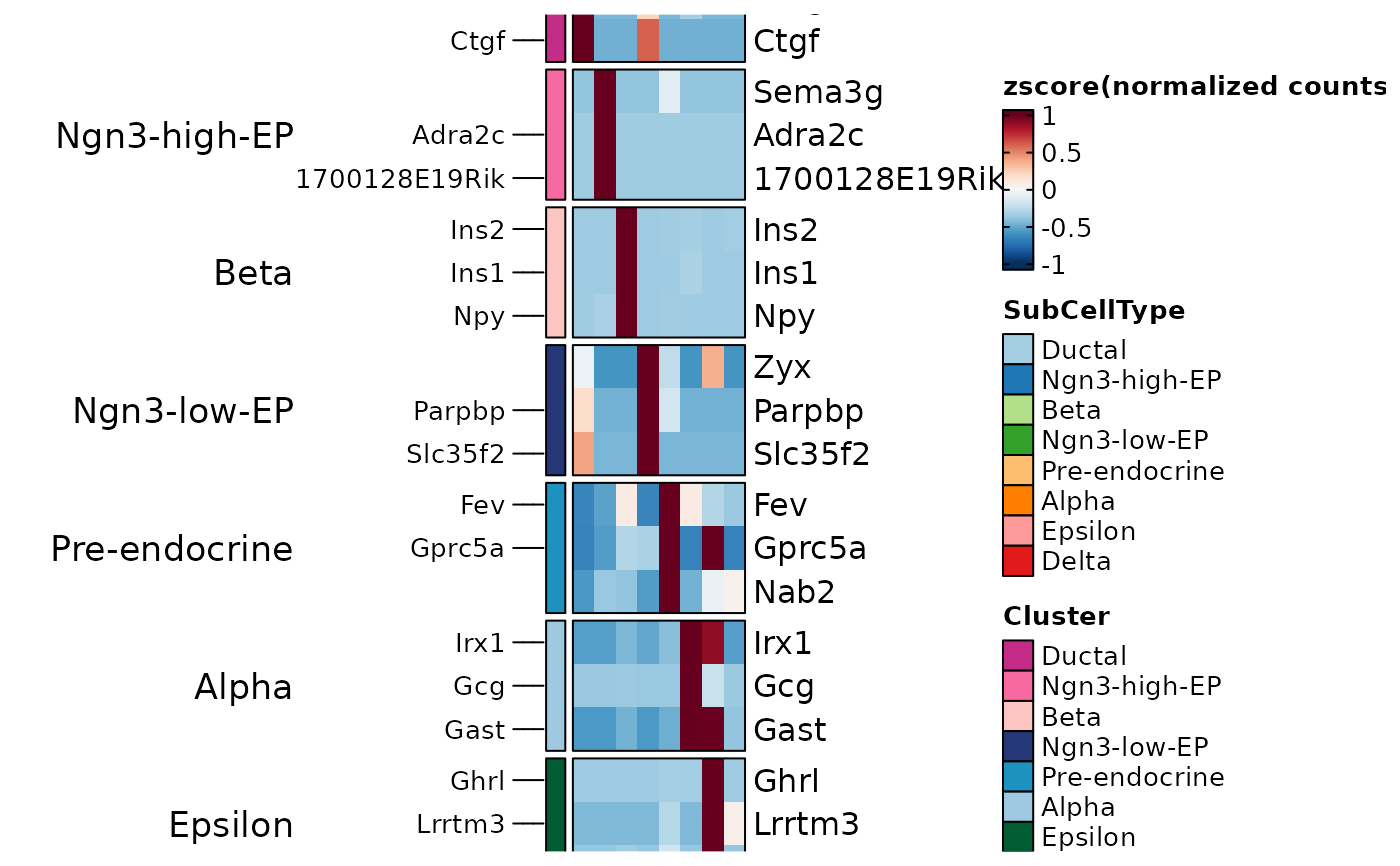
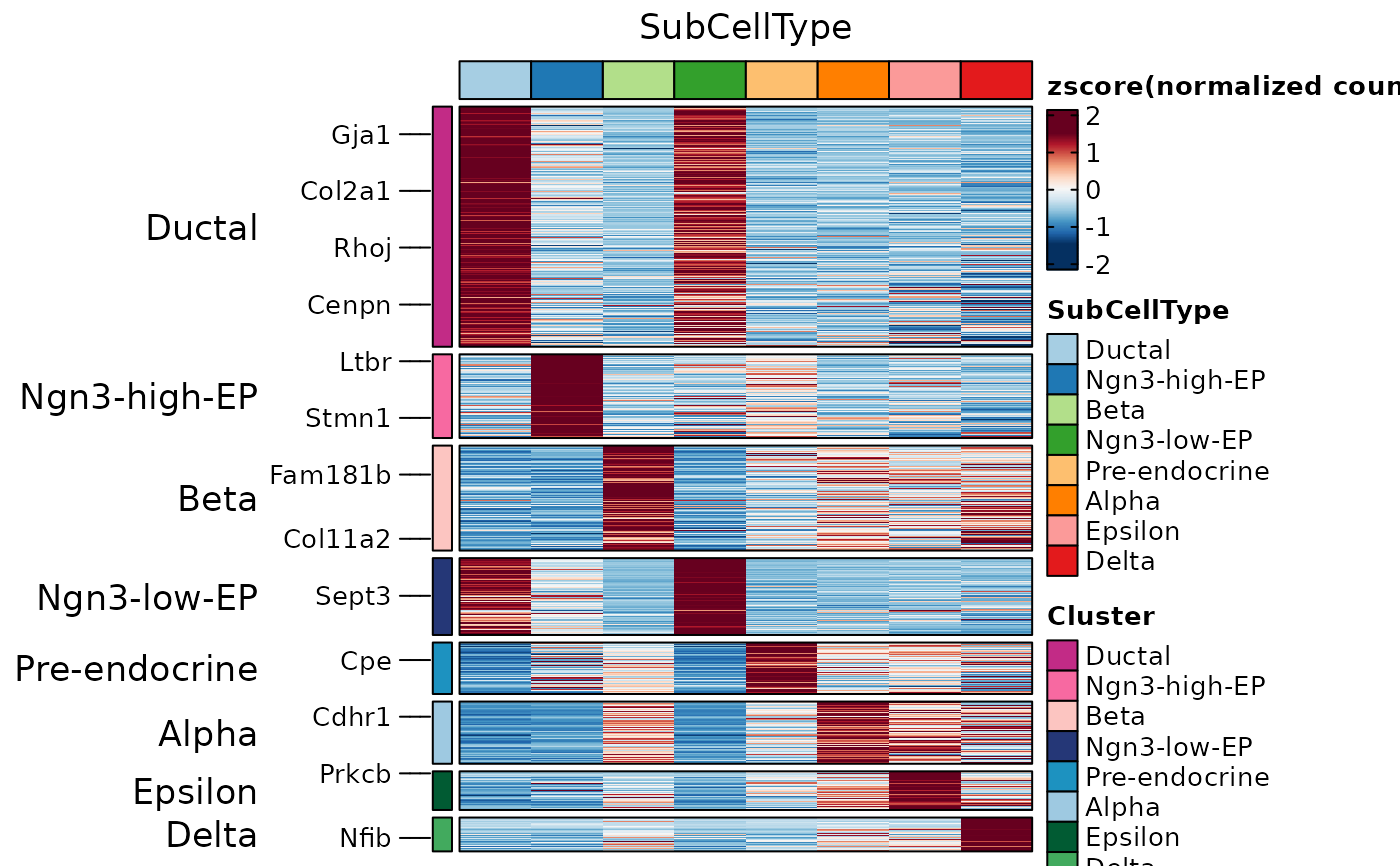 ht1$plot
ht1$plot
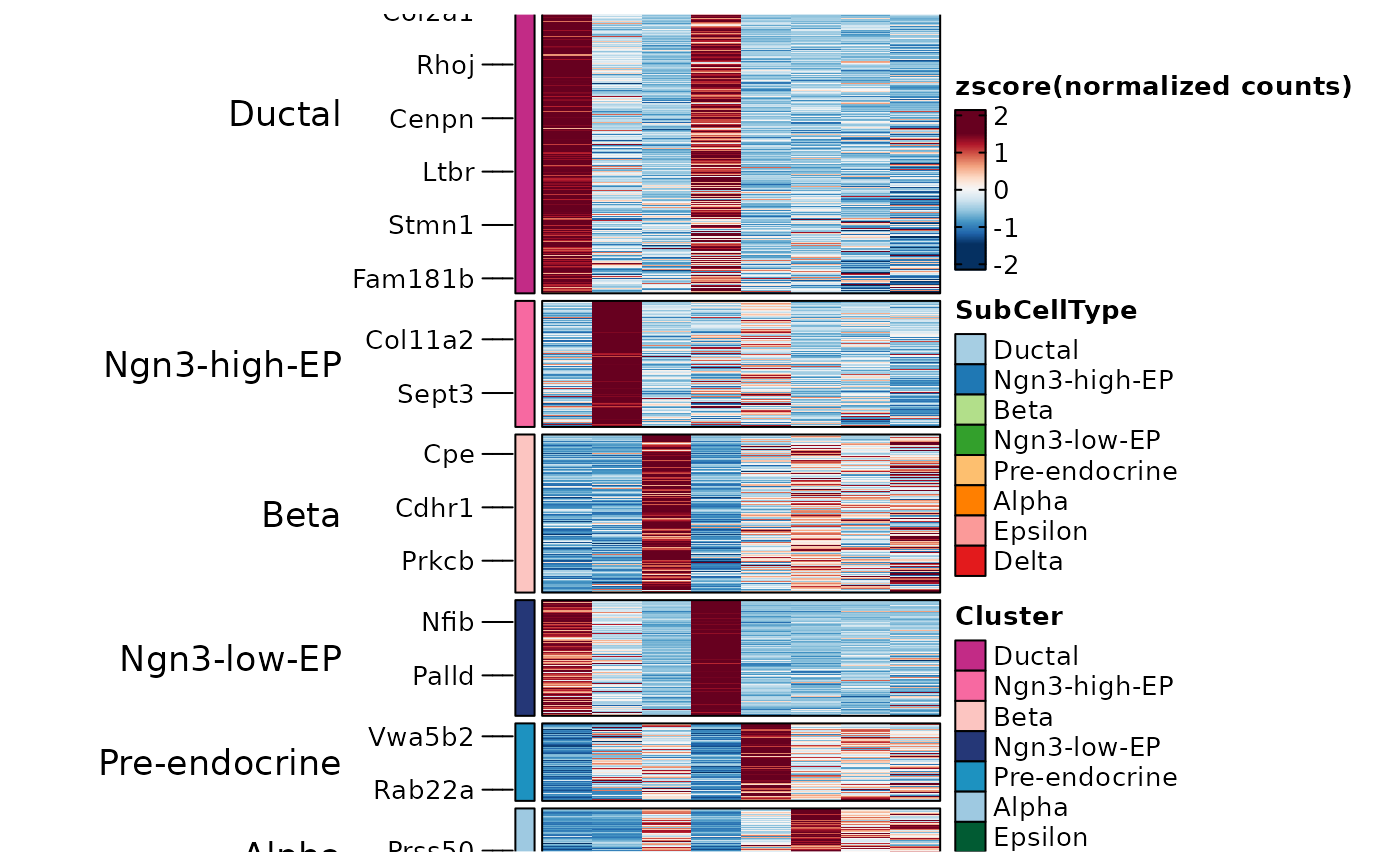 TopMarkers <- AllMarkers |>
dplyr::group_by(gene) |>
dplyr::top_n(1, avg_log2FC) |>
dplyr::group_by(group1) |>
dplyr::top_n(3, avg_log2FC)
ht2 <- GroupHeatmap(
pancreas_sub,
features = TopMarkers$gene,
feature_split = TopMarkers$group1,
group.by = "SubCellType",
show_row_names = TRUE
)
#> ℹ [2026-02-11 03:49:56] The size of the heatmap is fixed because certain elements are not scalable.
#> ℹ [2026-02-11 03:49:56] The width and height of the heatmap are determined by the size of the current viewport.
#> ℹ [2026-02-11 03:49:56] If you want to have more control over the size, you can manually set the parameters 'width' and 'height'.
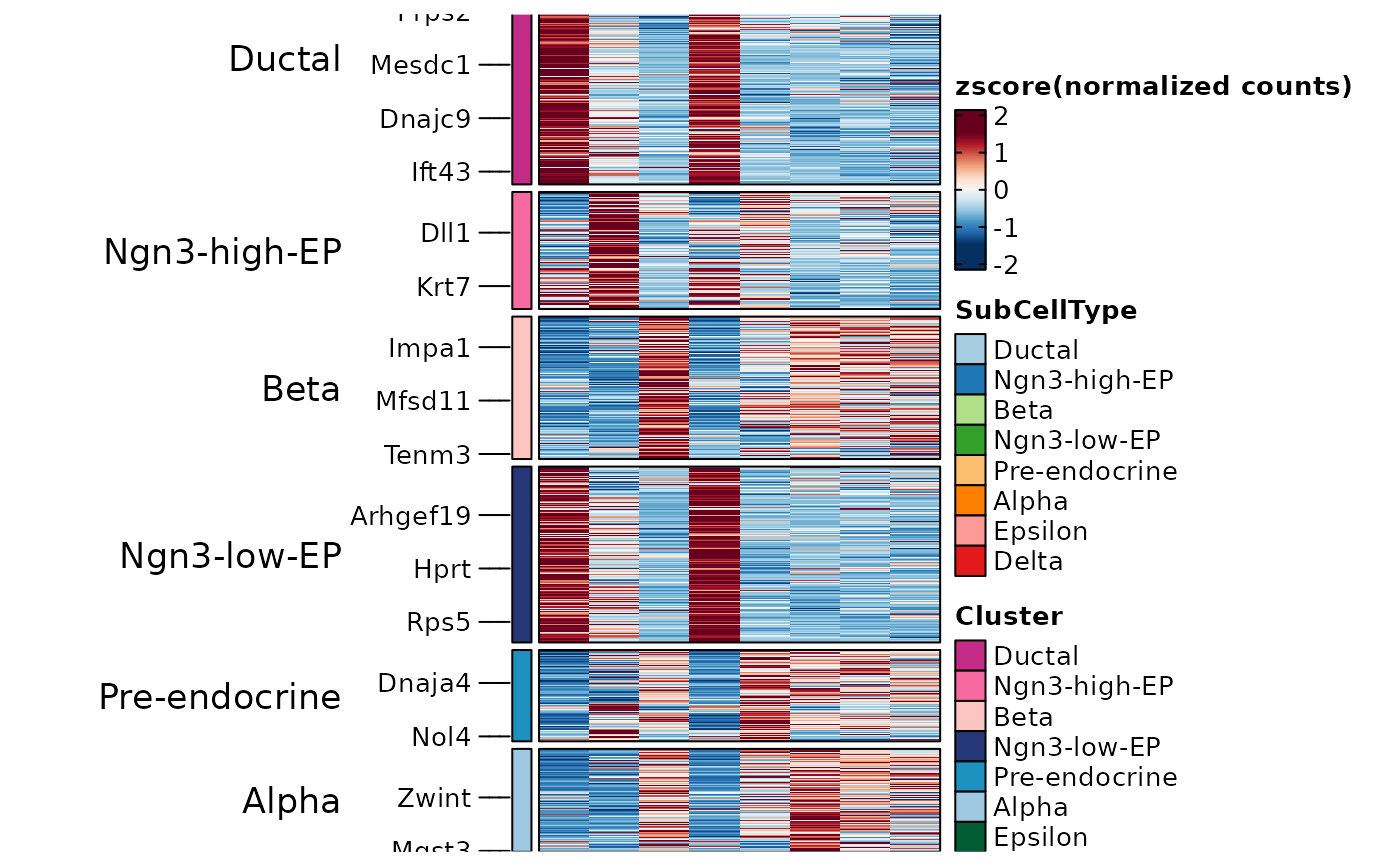
TopMarkers <- AllMarkers |>
dplyr::group_by(gene) |>
dplyr::top_n(1, avg_log2FC) |>
dplyr::group_by(group1) |>
dplyr::top_n(3, avg_log2FC)
ht2 <- GroupHeatmap(
pancreas_sub,
features = TopMarkers$gene,
feature_split = TopMarkers$group1,
group.by = "SubCellType",
show_row_names = TRUE
)
#> ℹ [2026-02-11 03:49:56] The size of the heatmap is fixed because certain elements are not scalable.
#> ℹ [2026-02-11 03:49:56] The width and height of the heatmap are determined by the size of the current viewport.
#> ℹ [2026-02-11 03:49:56] If you want to have more control over the size, you can manually set the parameters 'width' and 'height'.
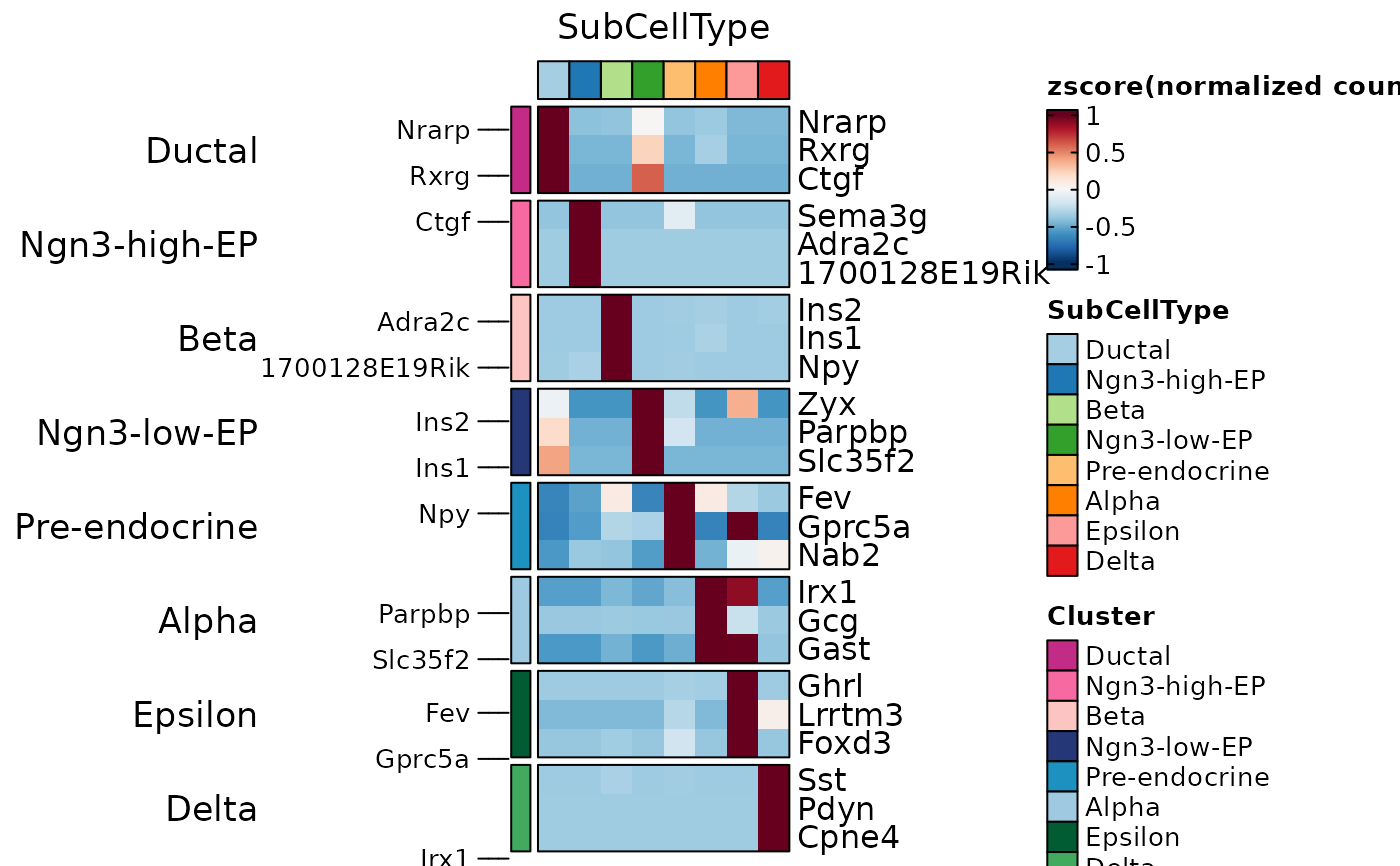 ht2$plot
ht2$plot
 pancreas_sub <- RunDEtest(
pancreas_sub,
group.by = "SubCellType",
markers_type = "paired",
cores = 2
)
#> ℹ [2026-02-11 03:49:57] Data type is log-normalized
#> ℹ [2026-02-11 03:49:57] Start differential expression test
#> ℹ [2026-02-11 03:49:57] Find paired markers(wilcox) among [1] 8 groups...
#> ℹ [2026-02-11 03:49:57] Using 2 cores
#> ⠙ [2026-02-11 03:49:57] Running for 1... [28/56] ■■■■■■■■■■■■■■■■ …
#> ✔ [2026-02-11 03:49:57] Completed 56 tasks in 4.9s
#>
#> ℹ [2026-02-11 03:49:57] Building results
#> ✔ [2026-02-11 03:50:02] Differential expression test completed
PairedMarkers <- dplyr::filter(
pancreas_sub@tools$DEtest_SubCellType$PairedMarkers_wilcox,
p_val_adj < 0.05 & avg_log2FC > 1
)
ht3 <- GroupHeatmap(
pancreas_sub,
features = PairedMarkers$gene,
feature_split = PairedMarkers$group1,
group.by = "SubCellType"
)
#> ℹ [2026-02-11 03:50:48] The size of the heatmap is fixed because certain elements are not scalable.
#> ℹ [2026-02-11 03:50:48] The width and height of the heatmap are determined by the size of the current viewport.
#> ℹ [2026-02-11 03:50:48] If you want to have more control over the size, you can manually set the parameters 'width' and 'height'.
pancreas_sub <- RunDEtest(
pancreas_sub,
group.by = "SubCellType",
markers_type = "paired",
cores = 2
)
#> ℹ [2026-02-11 03:49:57] Data type is log-normalized
#> ℹ [2026-02-11 03:49:57] Start differential expression test
#> ℹ [2026-02-11 03:49:57] Find paired markers(wilcox) among [1] 8 groups...
#> ℹ [2026-02-11 03:49:57] Using 2 cores
#> ⠙ [2026-02-11 03:49:57] Running for 1... [28/56] ■■■■■■■■■■■■■■■■ …
#> ✔ [2026-02-11 03:49:57] Completed 56 tasks in 4.9s
#>
#> ℹ [2026-02-11 03:49:57] Building results
#> ✔ [2026-02-11 03:50:02] Differential expression test completed
PairedMarkers <- dplyr::filter(
pancreas_sub@tools$DEtest_SubCellType$PairedMarkers_wilcox,
p_val_adj < 0.05 & avg_log2FC > 1
)
ht3 <- GroupHeatmap(
pancreas_sub,
features = PairedMarkers$gene,
feature_split = PairedMarkers$group1,
group.by = "SubCellType"
)
#> ℹ [2026-02-11 03:50:48] The size of the heatmap is fixed because certain elements are not scalable.
#> ℹ [2026-02-11 03:50:48] The width and height of the heatmap are determined by the size of the current viewport.
#> ℹ [2026-02-11 03:50:48] If you want to have more control over the size, you can manually set the parameters 'width' and 'height'.
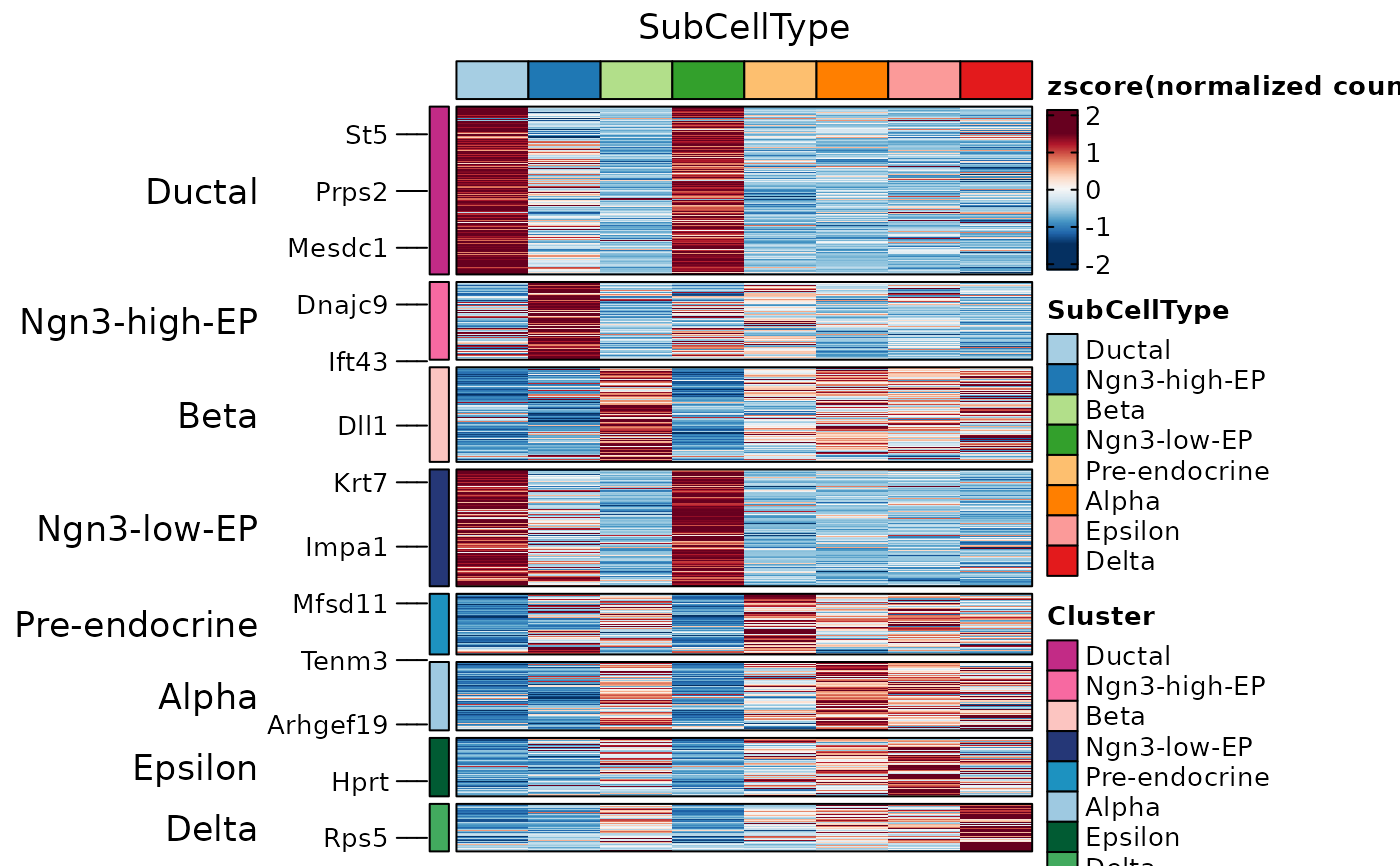 ht3$plot
ht3$plot
 data(panc8_sub)
panc8_sub <- integration_scop(
panc8_sub,
batch = "tech",
integration_method = "Uncorrected"
)
#> ◌ [2026-02-11 03:50:51] Run Uncorrected integration...
#> ℹ [2026-02-11 03:50:51] Spliting `srt_merge` into `srt_list` by column "tech"...
#> ℹ [2026-02-11 03:50:52] Checking a list of <Seurat>...
#> ! [2026-02-11 03:50:52] Data 1/5 of the `srt_list` is "unknown"
#> ℹ [2026-02-11 03:50:52] Perform `NormalizeData()` with `normalization.method = 'LogNormalize'` on the data 1/5 of the `srt_list`...
#> ℹ [2026-02-11 03:50:54] Perform `Seurat::FindVariableFeatures()` on the data 1/5 of the `srt_list`...
#> ! [2026-02-11 03:50:54] Data 2/5 of the `srt_list` is "unknown"
#> ℹ [2026-02-11 03:50:54] Perform `NormalizeData()` with `normalization.method = 'LogNormalize'` on the data 2/5 of the `srt_list`...
#> ℹ [2026-02-11 03:50:56] Perform `Seurat::FindVariableFeatures()` on the data 2/5 of the `srt_list`...
#> ! [2026-02-11 03:50:56] Data 3/5 of the `srt_list` is "unknown"
#> ℹ [2026-02-11 03:50:56] Perform `NormalizeData()` with `normalization.method = 'LogNormalize'` on the data 3/5 of the `srt_list`...
#> ℹ [2026-02-11 03:50:58] Perform `Seurat::FindVariableFeatures()` on the data 3/5 of the `srt_list`...
#> ! [2026-02-11 03:50:58] Data 4/5 of the `srt_list` is "unknown"
#> ℹ [2026-02-11 03:50:58] Perform `NormalizeData()` with `normalization.method = 'LogNormalize'` on the data 4/5 of the `srt_list`...
#> ℹ [2026-02-11 03:51:00] Perform `Seurat::FindVariableFeatures()` on the data 4/5 of the `srt_list`...
#> ! [2026-02-11 03:51:00] Data 5/5 of the `srt_list` is "unknown"
#> ℹ [2026-02-11 03:51:00] Perform `NormalizeData()` with `normalization.method = 'LogNormalize'` on the data 5/5 of the `srt_list`...
#> ℹ [2026-02-11 03:51:02] Perform `Seurat::FindVariableFeatures()` on the data 5/5 of the `srt_list`...
#> ℹ [2026-02-11 03:51:02] Use the separate HVF from srt_list
#> ℹ [2026-02-11 03:51:03] Number of available HVF: 2000
#> ℹ [2026-02-11 03:51:03] Finished check
#> ℹ [2026-02-11 03:51:05] Perform Uncorrected integration
#> ℹ [2026-02-11 03:51:06] Perform `Seurat::ScaleData()`
#> ℹ [2026-02-11 03:51:07] Perform linear dimension reduction("pca")
#> ℹ [2026-02-11 03:51:08] Perform Seurat::FindClusters ("louvain")
#> ℹ [2026-02-11 03:51:09] Reorder clusters...
#> ℹ [2026-02-11 03:51:09] Perform nonlinear dimension reduction ("umap")
#> ℹ [2026-02-11 03:51:09] Non-linear dimensionality reduction (umap) using (Uncorrectedpca) dims (1-10) as input
#> ℹ [2026-02-11 03:51:13] Non-linear dimensionality reduction (umap) using (Uncorrectedpca) dims (1-10) as input
#> ✔ [2026-02-11 03:51:20] Run Uncorrected integration done
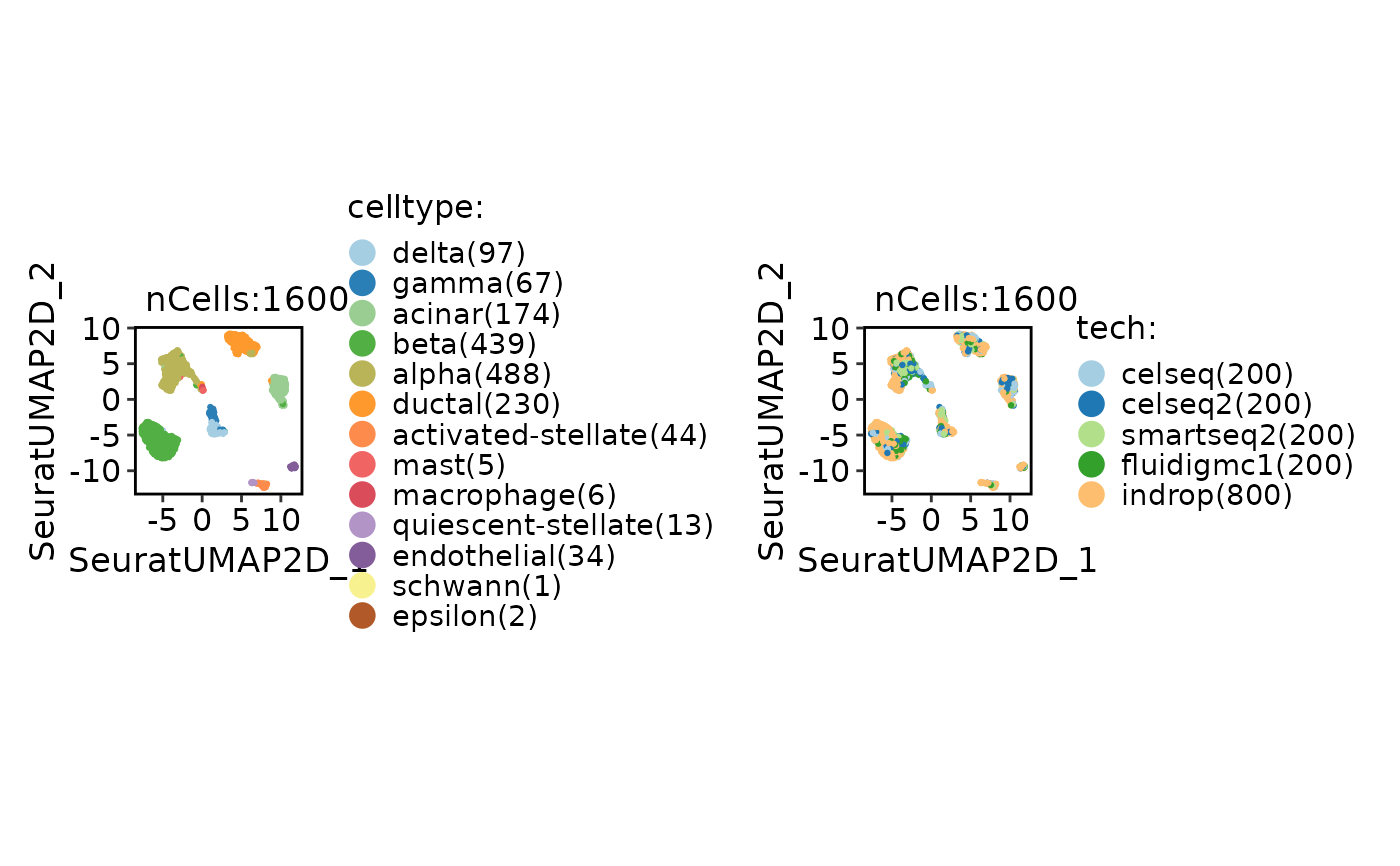
CellDimPlot(
panc8_sub,
group.by = c("celltype", "tech")
)
data(panc8_sub)
panc8_sub <- integration_scop(
panc8_sub,
batch = "tech",
integration_method = "Uncorrected"
)
#> ◌ [2026-02-11 03:50:51] Run Uncorrected integration...
#> ℹ [2026-02-11 03:50:51] Spliting `srt_merge` into `srt_list` by column "tech"...
#> ℹ [2026-02-11 03:50:52] Checking a list of <Seurat>...
#> ! [2026-02-11 03:50:52] Data 1/5 of the `srt_list` is "unknown"
#> ℹ [2026-02-11 03:50:52] Perform `NormalizeData()` with `normalization.method = 'LogNormalize'` on the data 1/5 of the `srt_list`...
#> ℹ [2026-02-11 03:50:54] Perform `Seurat::FindVariableFeatures()` on the data 1/5 of the `srt_list`...
#> ! [2026-02-11 03:50:54] Data 2/5 of the `srt_list` is "unknown"
#> ℹ [2026-02-11 03:50:54] Perform `NormalizeData()` with `normalization.method = 'LogNormalize'` on the data 2/5 of the `srt_list`...
#> ℹ [2026-02-11 03:50:56] Perform `Seurat::FindVariableFeatures()` on the data 2/5 of the `srt_list`...
#> ! [2026-02-11 03:50:56] Data 3/5 of the `srt_list` is "unknown"
#> ℹ [2026-02-11 03:50:56] Perform `NormalizeData()` with `normalization.method = 'LogNormalize'` on the data 3/5 of the `srt_list`...
#> ℹ [2026-02-11 03:50:58] Perform `Seurat::FindVariableFeatures()` on the data 3/5 of the `srt_list`...
#> ! [2026-02-11 03:50:58] Data 4/5 of the `srt_list` is "unknown"
#> ℹ [2026-02-11 03:50:58] Perform `NormalizeData()` with `normalization.method = 'LogNormalize'` on the data 4/5 of the `srt_list`...
#> ℹ [2026-02-11 03:51:00] Perform `Seurat::FindVariableFeatures()` on the data 4/5 of the `srt_list`...
#> ! [2026-02-11 03:51:00] Data 5/5 of the `srt_list` is "unknown"
#> ℹ [2026-02-11 03:51:00] Perform `NormalizeData()` with `normalization.method = 'LogNormalize'` on the data 5/5 of the `srt_list`...
#> ℹ [2026-02-11 03:51:02] Perform `Seurat::FindVariableFeatures()` on the data 5/5 of the `srt_list`...
#> ℹ [2026-02-11 03:51:02] Use the separate HVF from srt_list
#> ℹ [2026-02-11 03:51:03] Number of available HVF: 2000
#> ℹ [2026-02-11 03:51:03] Finished check
#> ℹ [2026-02-11 03:51:05] Perform Uncorrected integration
#> ℹ [2026-02-11 03:51:06] Perform `Seurat::ScaleData()`
#> ℹ [2026-02-11 03:51:07] Perform linear dimension reduction("pca")
#> ℹ [2026-02-11 03:51:08] Perform Seurat::FindClusters ("louvain")
#> ℹ [2026-02-11 03:51:09] Reorder clusters...
#> ℹ [2026-02-11 03:51:09] Perform nonlinear dimension reduction ("umap")
#> ℹ [2026-02-11 03:51:09] Non-linear dimensionality reduction (umap) using (Uncorrectedpca) dims (1-10) as input
#> ℹ [2026-02-11 03:51:13] Non-linear dimensionality reduction (umap) using (Uncorrectedpca) dims (1-10) as input
#> ✔ [2026-02-11 03:51:20] Run Uncorrected integration done
CellDimPlot(
panc8_sub,
group.by = c("celltype", "tech")
)
 panc8_sub <- RunDEtest(
srt = panc8_sub,
group.by = "celltype",
grouping.var = "tech",
markers_type = "conserved",
cores = 2
)
#> ℹ [2026-02-11 03:51:21] Data type is log-normalized
#> ℹ [2026-02-11 03:51:21] Start differential expression test
#> ℹ [2026-02-11 03:51:21] Find conserved markers(wilcox) among [1] 13 groups...
#> ℹ [2026-02-11 03:51:21] Using 2 cores
#> ⠙ [2026-02-11 03:51:21] Running for delta... [7/13] ■■■■■■■■■■■■■■■■■ …
#> ✔ [2026-02-11 03:51:21] Completed 13 tasks in 6.8s
#>
#> ℹ [2026-02-11 03:51:21] Building results
#> ✔ [2026-02-11 03:51:28] Differential expression test completed
ConservedMarkers1 <- dplyr::filter(
panc8_sub@tools$DEtest_celltype$ConservedMarkers_wilcox,
p_val_adj < 0.05 & avg_log2FC > 1
)
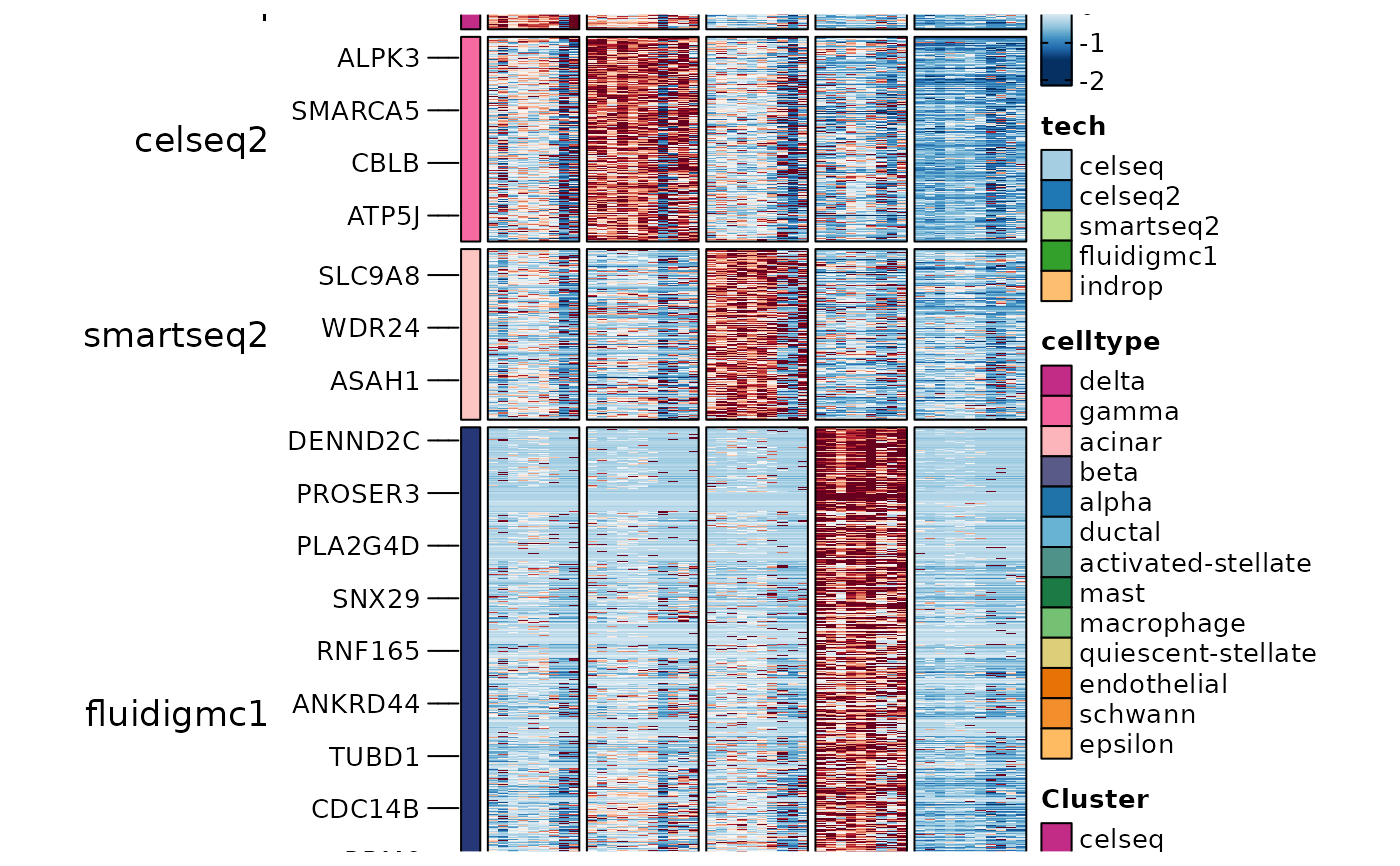
ht4 <- GroupHeatmap(
panc8_sub,
layer = "data",
features = ConservedMarkers1$gene,
feature_split = ConservedMarkers1$group1,
group.by = "tech",
split.by = "celltype",
within_groups = TRUE
)
#> `use_raster` is automatically set to TRUE for a matrix with more than
#> 2000 rows. You can control `use_raster` argument by explicitly setting
#> TRUE/FALSE to it.
#>
#> Set `ht_opt$message = FALSE` to turn off this message.
#> `use_raster` is automatically set to TRUE for a matrix with more than
#> 2000 rows. You can control `use_raster` argument by explicitly setting
#> TRUE/FALSE to it.
#>
#> Set `ht_opt$message = FALSE` to turn off this message.
#> ℹ [2026-02-11 03:51:38] The size of the heatmap is fixed because certain elements are not scalable.
#> ℹ [2026-02-11 03:51:38] The width and height of the heatmap are determined by the size of the current viewport.
#> ℹ [2026-02-11 03:51:38] If you want to have more control over the size, you can manually set the parameters 'width' and 'height'.
panc8_sub <- RunDEtest(
srt = panc8_sub,
group.by = "celltype",
grouping.var = "tech",
markers_type = "conserved",
cores = 2
)
#> ℹ [2026-02-11 03:51:21] Data type is log-normalized
#> ℹ [2026-02-11 03:51:21] Start differential expression test
#> ℹ [2026-02-11 03:51:21] Find conserved markers(wilcox) among [1] 13 groups...
#> ℹ [2026-02-11 03:51:21] Using 2 cores
#> ⠙ [2026-02-11 03:51:21] Running for delta... [7/13] ■■■■■■■■■■■■■■■■■ …
#> ✔ [2026-02-11 03:51:21] Completed 13 tasks in 6.8s
#>
#> ℹ [2026-02-11 03:51:21] Building results
#> ✔ [2026-02-11 03:51:28] Differential expression test completed
ConservedMarkers1 <- dplyr::filter(
panc8_sub@tools$DEtest_celltype$ConservedMarkers_wilcox,
p_val_adj < 0.05 & avg_log2FC > 1
)
ht4 <- GroupHeatmap(
panc8_sub,
layer = "data",
features = ConservedMarkers1$gene,
feature_split = ConservedMarkers1$group1,
group.by = "tech",
split.by = "celltype",
within_groups = TRUE
)
#> `use_raster` is automatically set to TRUE for a matrix with more than
#> 2000 rows. You can control `use_raster` argument by explicitly setting
#> TRUE/FALSE to it.
#>
#> Set `ht_opt$message = FALSE` to turn off this message.
#> `use_raster` is automatically set to TRUE for a matrix with more than
#> 2000 rows. You can control `use_raster` argument by explicitly setting
#> TRUE/FALSE to it.
#>
#> Set `ht_opt$message = FALSE` to turn off this message.
#> ℹ [2026-02-11 03:51:38] The size of the heatmap is fixed because certain elements are not scalable.
#> ℹ [2026-02-11 03:51:38] The width and height of the heatmap are determined by the size of the current viewport.
#> ℹ [2026-02-11 03:51:38] If you want to have more control over the size, you can manually set the parameters 'width' and 'height'.
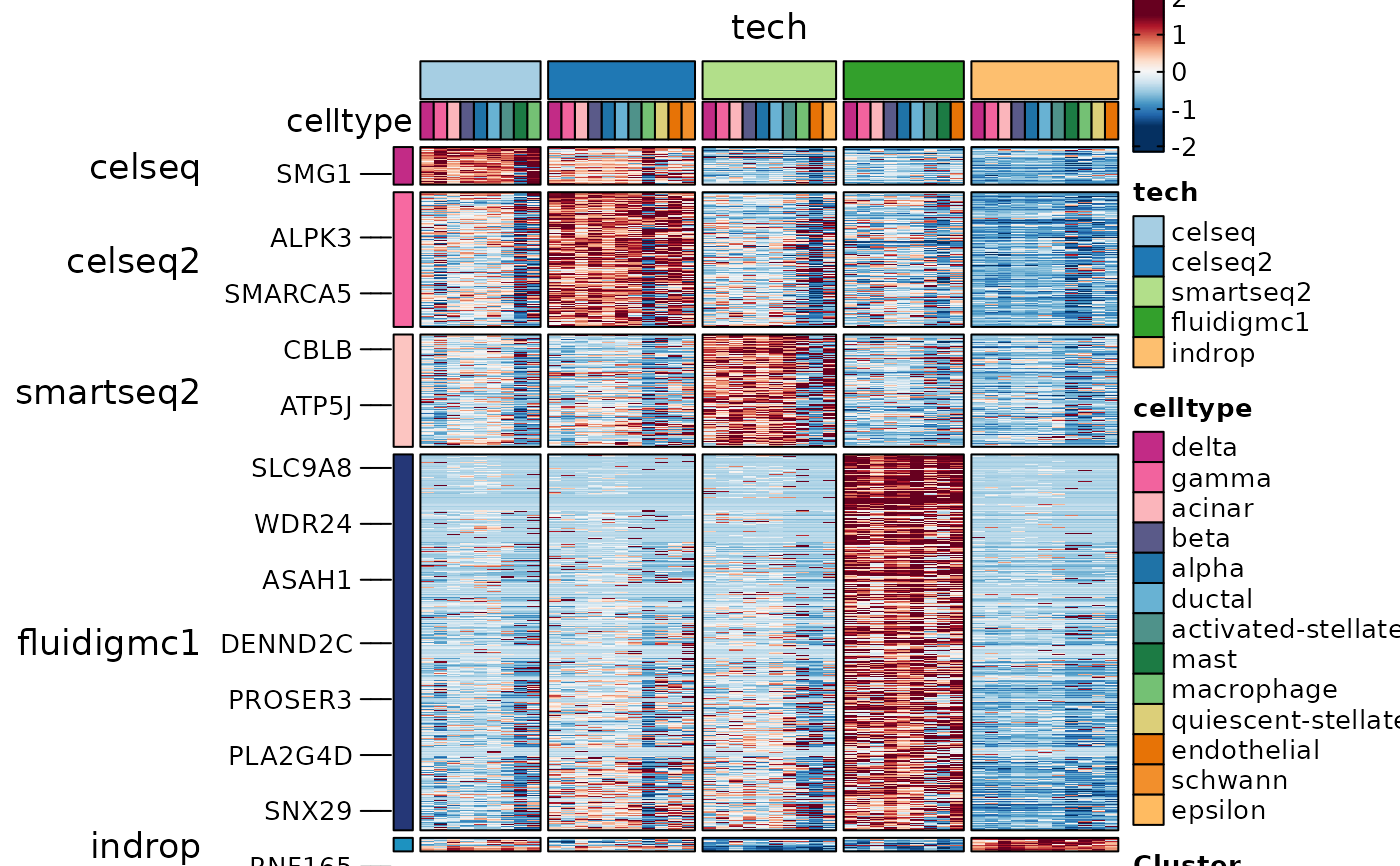 ht4$plot
ht4$plot
 panc8_sub <- RunDEtest(
srt = panc8_sub,
group.by = "tech",
grouping.var = "celltype",
markers_type = "conserved",
cores = 2
)
#> ℹ [2026-02-11 03:51:48] Data type is log-normalized
#> ℹ [2026-02-11 03:51:48] Start differential expression test
#> ℹ [2026-02-11 03:51:48] Find conserved markers(wilcox) among [1] 5 groups...
#> ℹ [2026-02-11 03:51:48] Using 2 cores
#> ⠙ [2026-02-11 03:51:48] Running for celseq... [3/5] ■■■■■■■■■■■■■■■■■■■ …
#> ✔ [2026-02-11 03:51:48] Completed 5 tasks in 6.4s
#>
#> ℹ [2026-02-11 03:51:48] Building results
#> ✔ [2026-02-11 03:51:55] Differential expression test completed
ConservedMarkers2 <- dplyr::filter(
panc8_sub@tools$DEtest_tech$ConservedMarkers_wilcox,
p_val_adj < 0.05 & avg_log2FC > 1
)
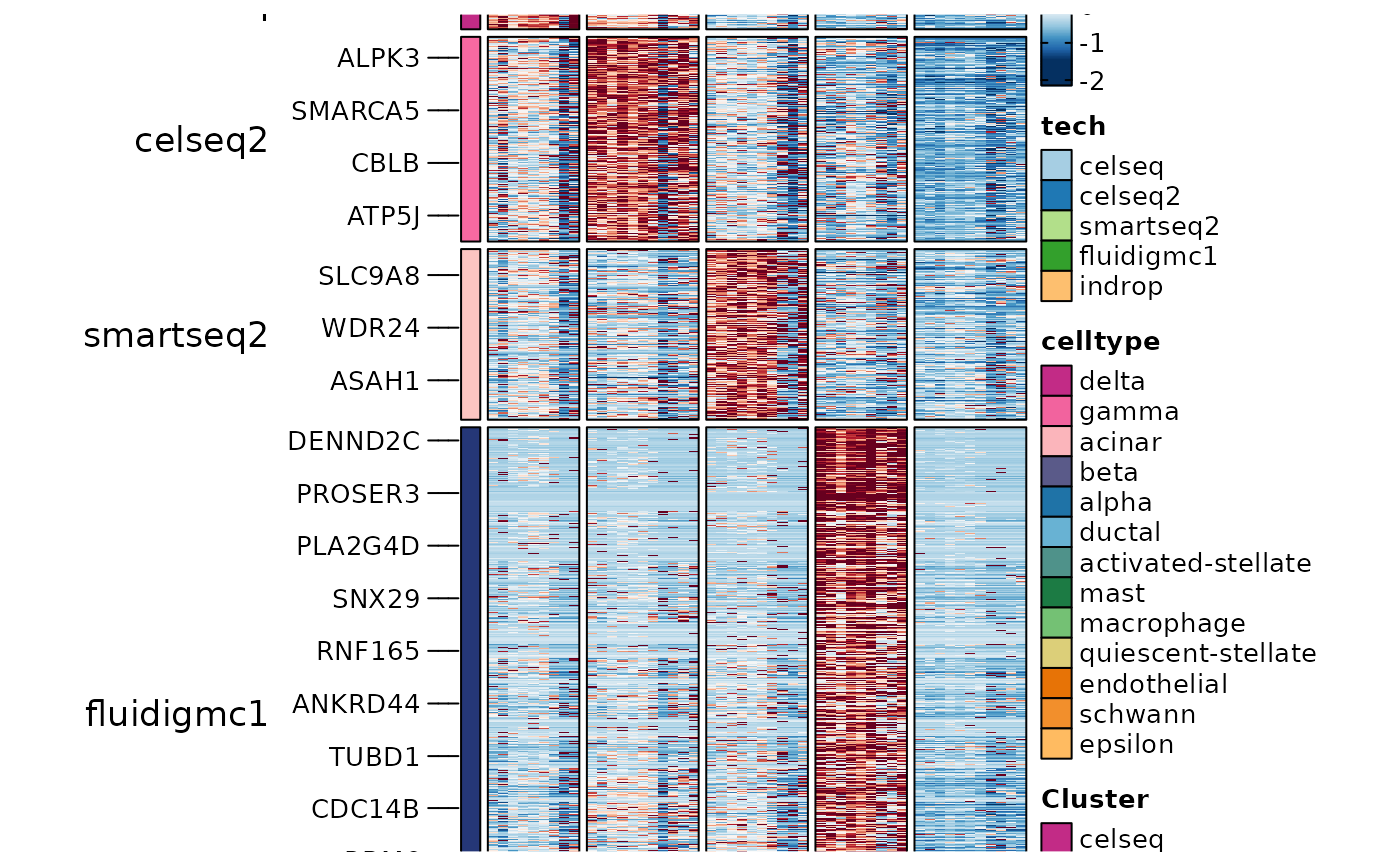
ht4 <- GroupHeatmap(
srt = panc8_sub,
layer = "data",
features = ConservedMarkers2$gene,
feature_split = ConservedMarkers2$group1,
group.by = "tech",
split.by = "celltype"
)
#> ℹ [2026-02-11 03:51:58] The size of the heatmap is fixed because certain elements are not scalable.
#> ℹ [2026-02-11 03:51:58] The width and height of the heatmap are determined by the size of the current viewport.
#> ℹ [2026-02-11 03:51:58] If you want to have more control over the size, you can manually set the parameters 'width' and 'height'.
panc8_sub <- RunDEtest(
srt = panc8_sub,
group.by = "tech",
grouping.var = "celltype",
markers_type = "conserved",
cores = 2
)
#> ℹ [2026-02-11 03:51:48] Data type is log-normalized
#> ℹ [2026-02-11 03:51:48] Start differential expression test
#> ℹ [2026-02-11 03:51:48] Find conserved markers(wilcox) among [1] 5 groups...
#> ℹ [2026-02-11 03:51:48] Using 2 cores
#> ⠙ [2026-02-11 03:51:48] Running for celseq... [3/5] ■■■■■■■■■■■■■■■■■■■ …
#> ✔ [2026-02-11 03:51:48] Completed 5 tasks in 6.4s
#>
#> ℹ [2026-02-11 03:51:48] Building results
#> ✔ [2026-02-11 03:51:55] Differential expression test completed
ConservedMarkers2 <- dplyr::filter(
panc8_sub@tools$DEtest_tech$ConservedMarkers_wilcox,
p_val_adj < 0.05 & avg_log2FC > 1
)
ht4 <- GroupHeatmap(
srt = panc8_sub,
layer = "data",
features = ConservedMarkers2$gene,
feature_split = ConservedMarkers2$group1,
group.by = "tech",
split.by = "celltype"
)
#> ℹ [2026-02-11 03:51:58] The size of the heatmap is fixed because certain elements are not scalable.
#> ℹ [2026-02-11 03:51:58] The width and height of the heatmap are determined by the size of the current viewport.
#> ℹ [2026-02-11 03:51:58] If you want to have more control over the size, you can manually set the parameters 'width' and 'height'.
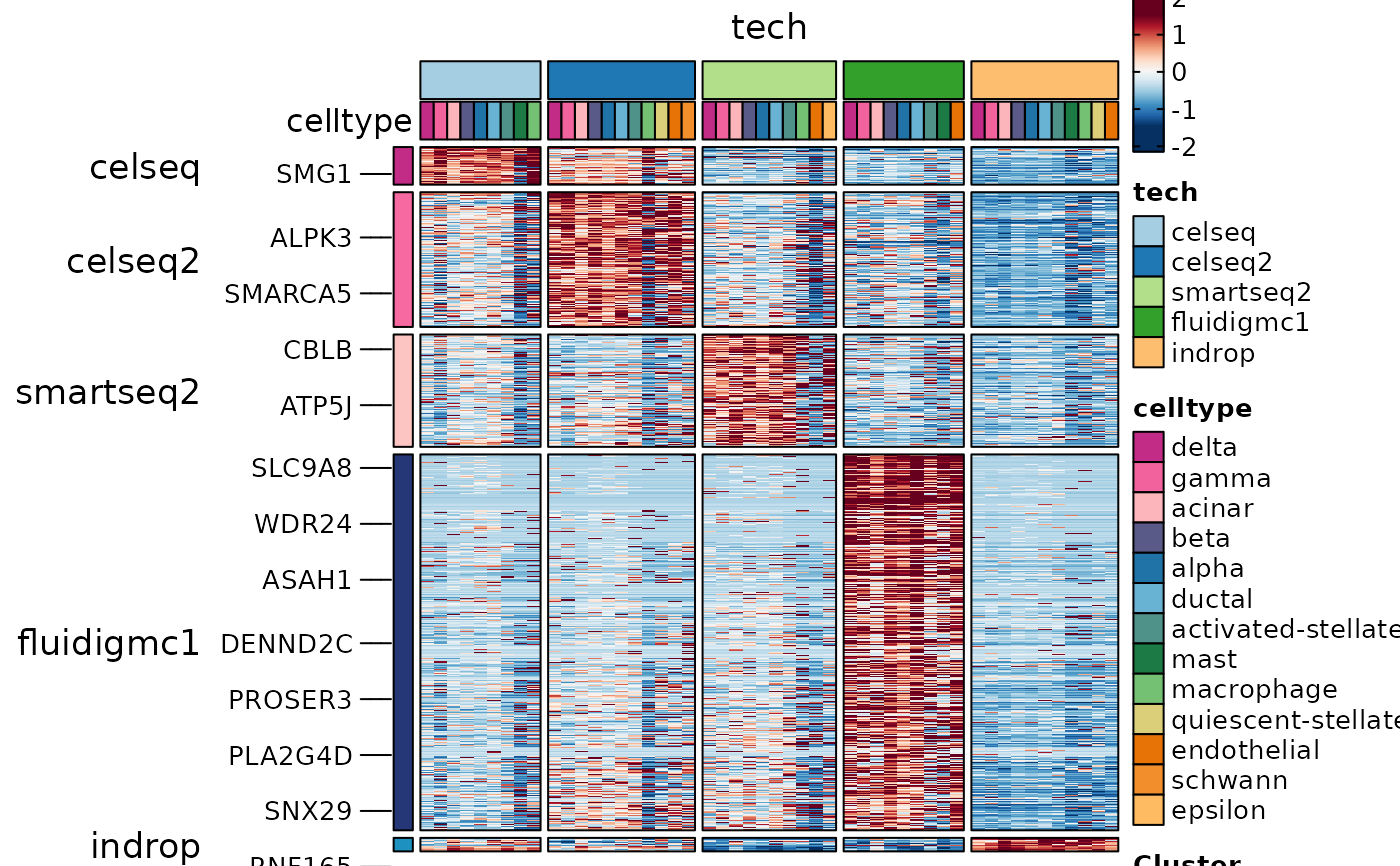 ht4$plot
ht4$plot
 panc8_sub <- RunDEtest(
srt = panc8_sub,
group.by = "celltype",
grouping.var = "tech",
markers_type = "disturbed",
cores = 2
)
#> ℹ [2026-02-11 03:52:03] Data type is log-normalized
#> ℹ [2026-02-11 03:52:03] Start differential expression test
#> ℹ [2026-02-11 03:52:03] Find disturbed markers(wilcox) among [1] 13 groups...
#> ℹ [2026-02-11 03:52:03] Using 2 cores
#> ⠙ [2026-02-11 03:52:03] Running for delta... [7/13] ■■■■■■■■■■■■■■■■■ …
#> ✔ [2026-02-11 03:52:03] Completed 13 tasks in 13.4s
#>
#> ℹ [2026-02-11 03:52:03] Building results
#> ✔ [2026-02-11 03:52:17] Differential expression test completed
DisturbedMarkers <- dplyr::filter(
panc8_sub@tools$DEtest_celltype$DisturbedMarkers_wilcox,
p_val_adj < 0.05 & avg_log2FC > 1 & var1 == "smartseq2"
)
ht5 <- GroupHeatmap(
srt = panc8_sub,
layer = "data",
features = DisturbedMarkers$gene,
feature_split = DisturbedMarkers$group1,
group.by = "celltype",
split.by = "tech"
)
#> `use_raster` is automatically set to TRUE for a matrix with more than
#> 2000 rows. You can control `use_raster` argument by explicitly setting
#> TRUE/FALSE to it.
#>
#> Set `ht_opt$message = FALSE` to turn off this message.
#> ℹ [2026-02-11 03:52:28] The size of the heatmap is fixed because certain elements are not scalable.
#> ℹ [2026-02-11 03:52:28] The width and height of the heatmap are determined by the size of the current viewport.
#> ℹ [2026-02-11 03:52:28] If you want to have more control over the size, you can manually set the parameters 'width' and 'height'.
panc8_sub <- RunDEtest(
srt = panc8_sub,
group.by = "celltype",
grouping.var = "tech",
markers_type = "disturbed",
cores = 2
)
#> ℹ [2026-02-11 03:52:03] Data type is log-normalized
#> ℹ [2026-02-11 03:52:03] Start differential expression test
#> ℹ [2026-02-11 03:52:03] Find disturbed markers(wilcox) among [1] 13 groups...
#> ℹ [2026-02-11 03:52:03] Using 2 cores
#> ⠙ [2026-02-11 03:52:03] Running for delta... [7/13] ■■■■■■■■■■■■■■■■■ …
#> ✔ [2026-02-11 03:52:03] Completed 13 tasks in 13.4s
#>
#> ℹ [2026-02-11 03:52:03] Building results
#> ✔ [2026-02-11 03:52:17] Differential expression test completed
DisturbedMarkers <- dplyr::filter(
panc8_sub@tools$DEtest_celltype$DisturbedMarkers_wilcox,
p_val_adj < 0.05 & avg_log2FC > 1 & var1 == "smartseq2"
)
ht5 <- GroupHeatmap(
srt = panc8_sub,
layer = "data",
features = DisturbedMarkers$gene,
feature_split = DisturbedMarkers$group1,
group.by = "celltype",
split.by = "tech"
)
#> `use_raster` is automatically set to TRUE for a matrix with more than
#> 2000 rows. You can control `use_raster` argument by explicitly setting
#> TRUE/FALSE to it.
#>
#> Set `ht_opt$message = FALSE` to turn off this message.
#> ℹ [2026-02-11 03:52:28] The size of the heatmap is fixed because certain elements are not scalable.
#> ℹ [2026-02-11 03:52:28] The width and height of the heatmap are determined by the size of the current viewport.
#> ℹ [2026-02-11 03:52:28] If you want to have more control over the size, you can manually set the parameters 'width' and 'height'.
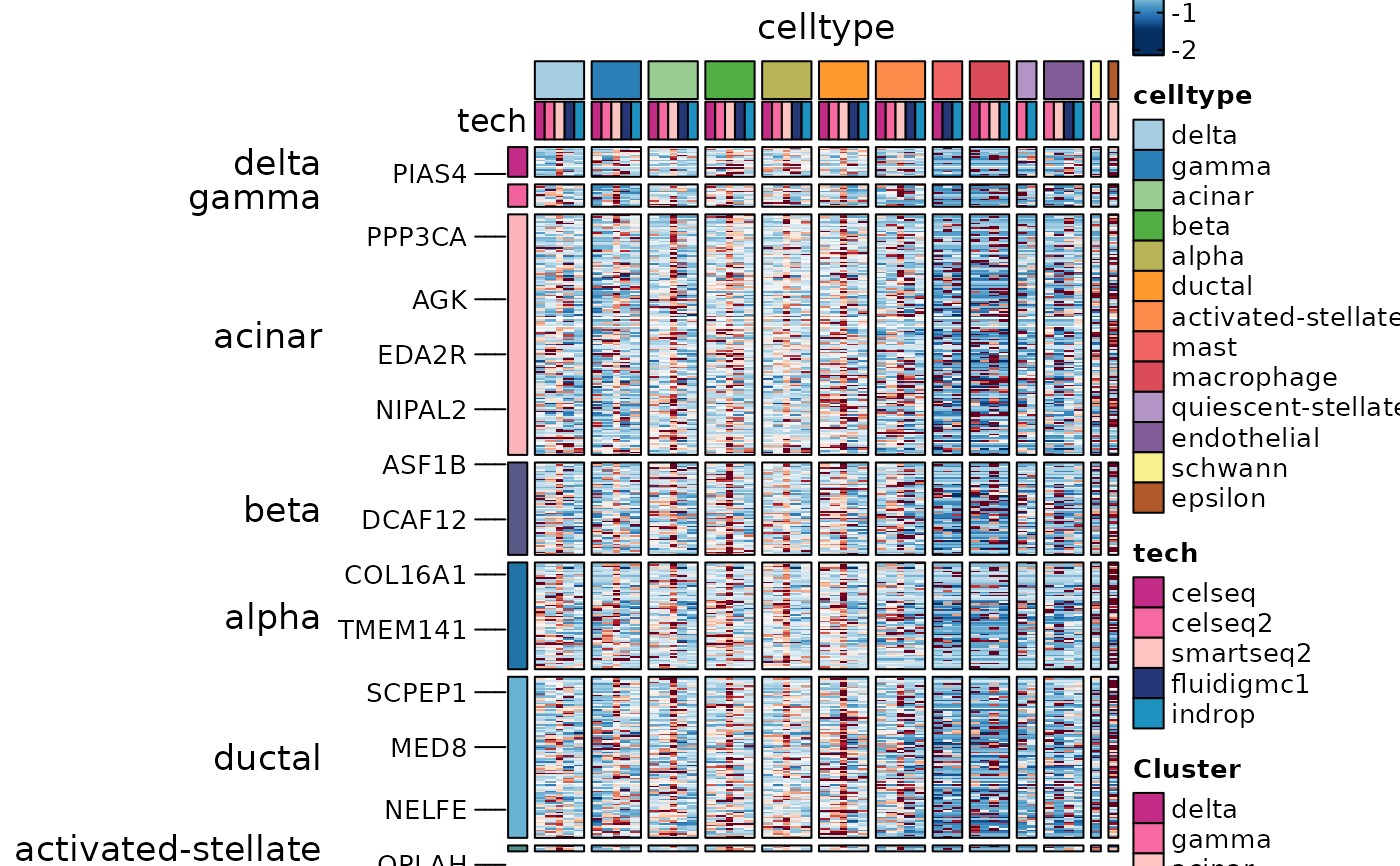 ht5$plot
ht5$plot
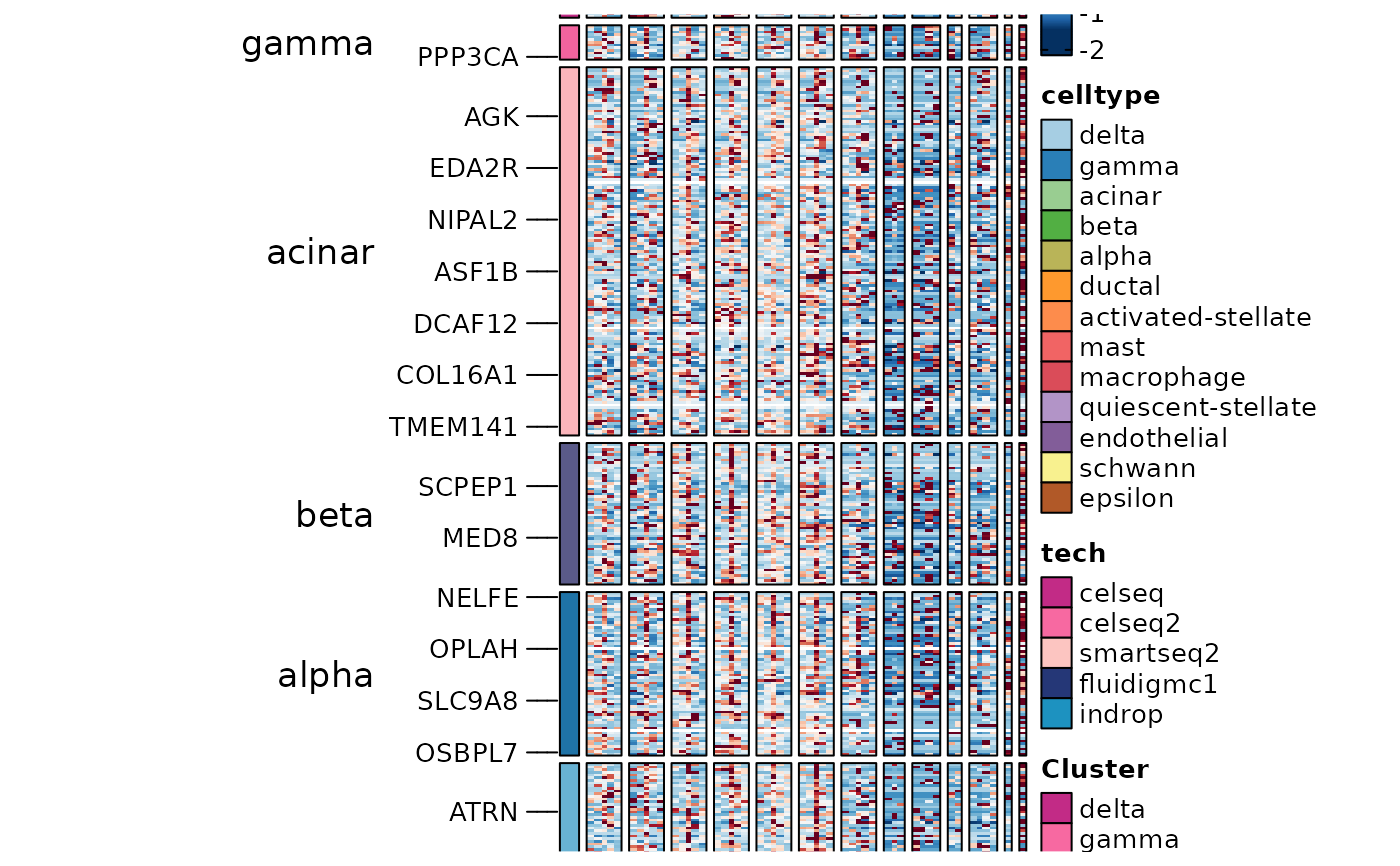 gene_specific <- names(which(table(DisturbedMarkers$gene) == 1))
DisturbedMarkers_specific <- DisturbedMarkers[
DisturbedMarkers$gene %in% gene_specific,
]
ht6 <- GroupHeatmap(
srt = panc8_sub,
layer = "data",
features = DisturbedMarkers_specific$gene,
feature_split = DisturbedMarkers_specific$group1,
group.by = "celltype",
split.by = "tech"
)
#> ℹ [2026-02-11 03:52:46] The size of the heatmap is fixed because certain elements are not scalable.
#> ℹ [2026-02-11 03:52:46] The width and height of the heatmap are determined by the size of the current viewport.
#> ℹ [2026-02-11 03:52:46] If you want to have more control over the size, you can manually set the parameters 'width' and 'height'.
gene_specific <- names(which(table(DisturbedMarkers$gene) == 1))
DisturbedMarkers_specific <- DisturbedMarkers[
DisturbedMarkers$gene %in% gene_specific,
]
ht6 <- GroupHeatmap(
srt = panc8_sub,
layer = "data",
features = DisturbedMarkers_specific$gene,
feature_split = DisturbedMarkers_specific$group1,
group.by = "celltype",
split.by = "tech"
)
#> ℹ [2026-02-11 03:52:46] The size of the heatmap is fixed because certain elements are not scalable.
#> ℹ [2026-02-11 03:52:46] The width and height of the heatmap are determined by the size of the current viewport.
#> ℹ [2026-02-11 03:52:46] If you want to have more control over the size, you can manually set the parameters 'width' and 'height'.
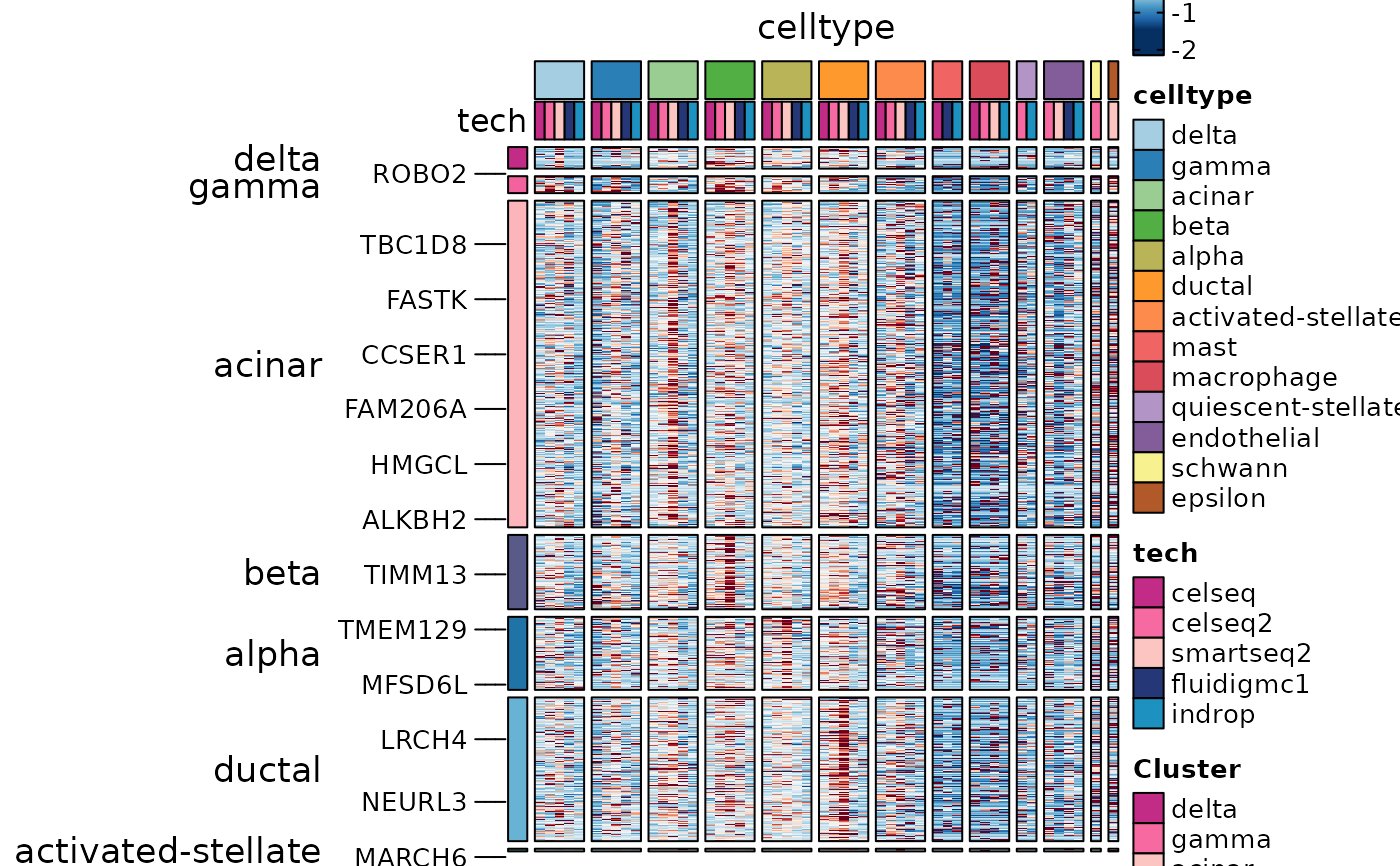 ht6$plot
ht6$plot
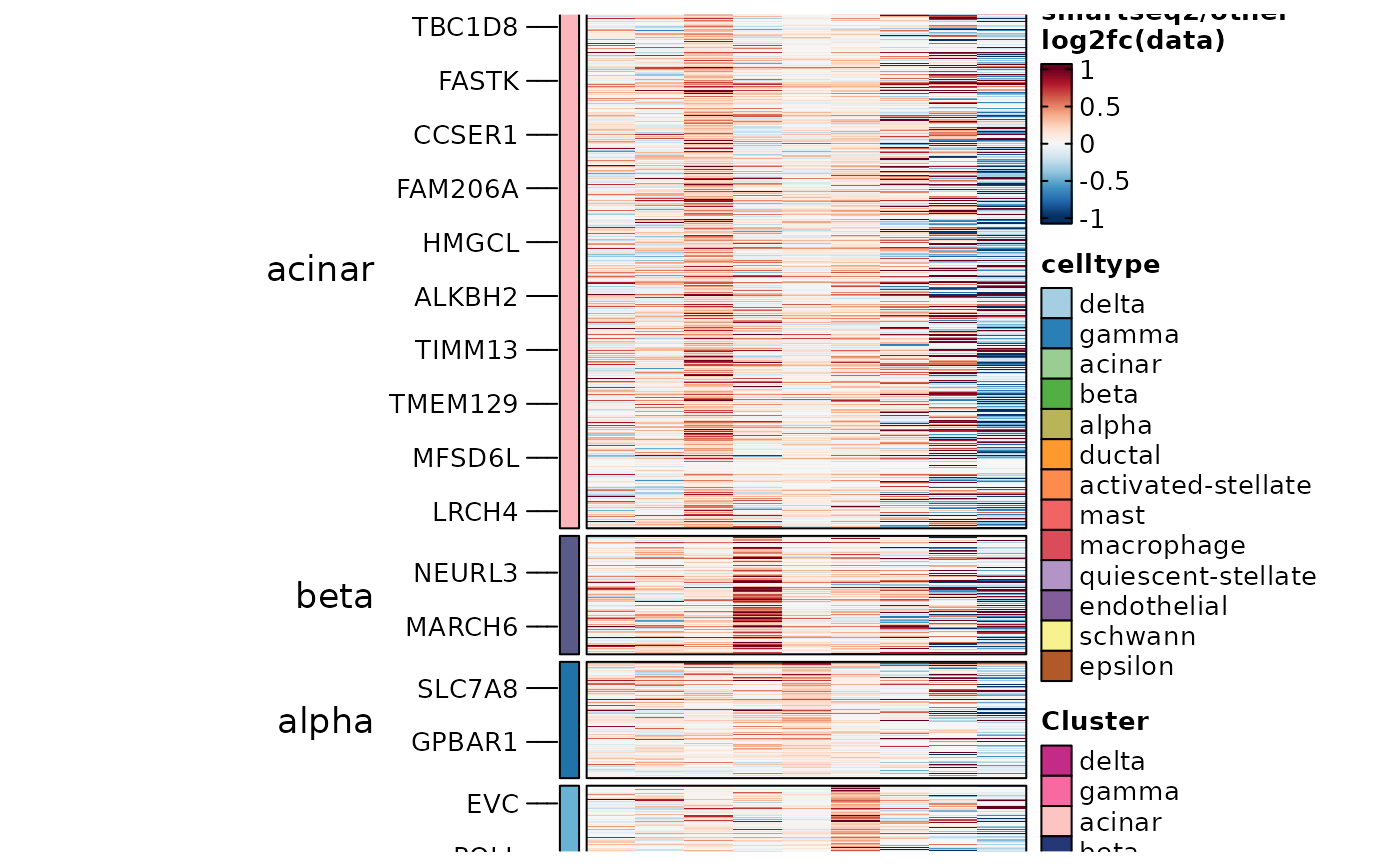
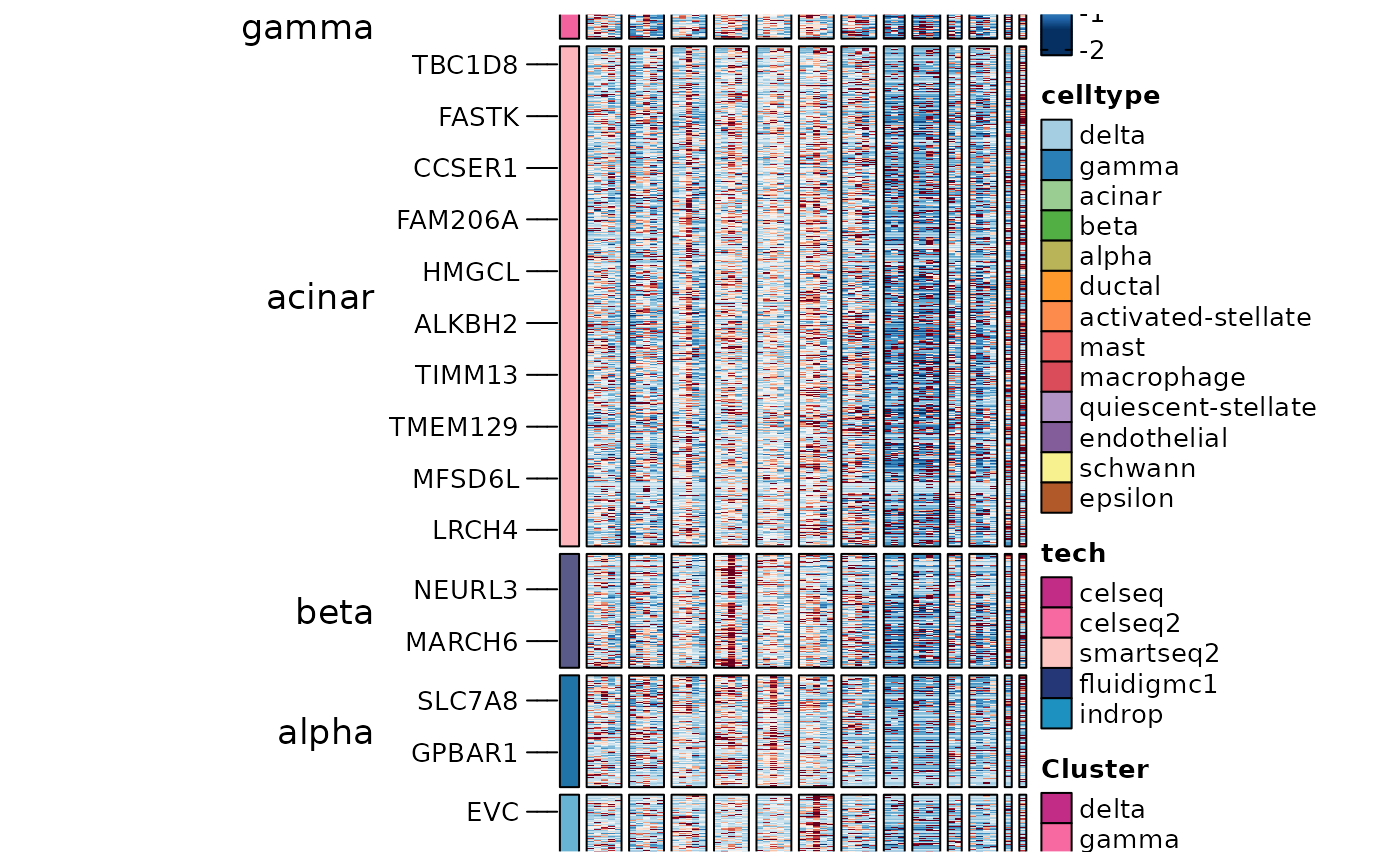 ht7 <- GroupHeatmap(
srt = panc8_sub,
layer = "data",
aggregate_fun = function(x) mean(expm1(x)) + 1,
features = DisturbedMarkers_specific$gene,
feature_split = DisturbedMarkers_specific$group1,
group.by = "celltype",
grouping.var = "tech",
numerator = "smartseq2"
)
#> ! [2026-02-11 03:52:57] When 'grouping.var' is specified, 'exp_method' can only be 'log2fc'
#> ℹ [2026-02-11 03:53:00] The size of the heatmap is fixed because certain elements are not scalable.
#> ℹ [2026-02-11 03:53:00] The width and height of the heatmap are determined by the size of the current viewport.
#> ℹ [2026-02-11 03:53:00] If you want to have more control over the size, you can manually set the parameters 'width' and 'height'.
ht7 <- GroupHeatmap(
srt = panc8_sub,
layer = "data",
aggregate_fun = function(x) mean(expm1(x)) + 1,
features = DisturbedMarkers_specific$gene,
feature_split = DisturbedMarkers_specific$group1,
group.by = "celltype",
grouping.var = "tech",
numerator = "smartseq2"
)
#> ! [2026-02-11 03:52:57] When 'grouping.var' is specified, 'exp_method' can only be 'log2fc'
#> ℹ [2026-02-11 03:53:00] The size of the heatmap is fixed because certain elements are not scalable.
#> ℹ [2026-02-11 03:53:00] The width and height of the heatmap are determined by the size of the current viewport.
#> ℹ [2026-02-11 03:53:00] If you want to have more control over the size, you can manually set the parameters 'width' and 'height'.
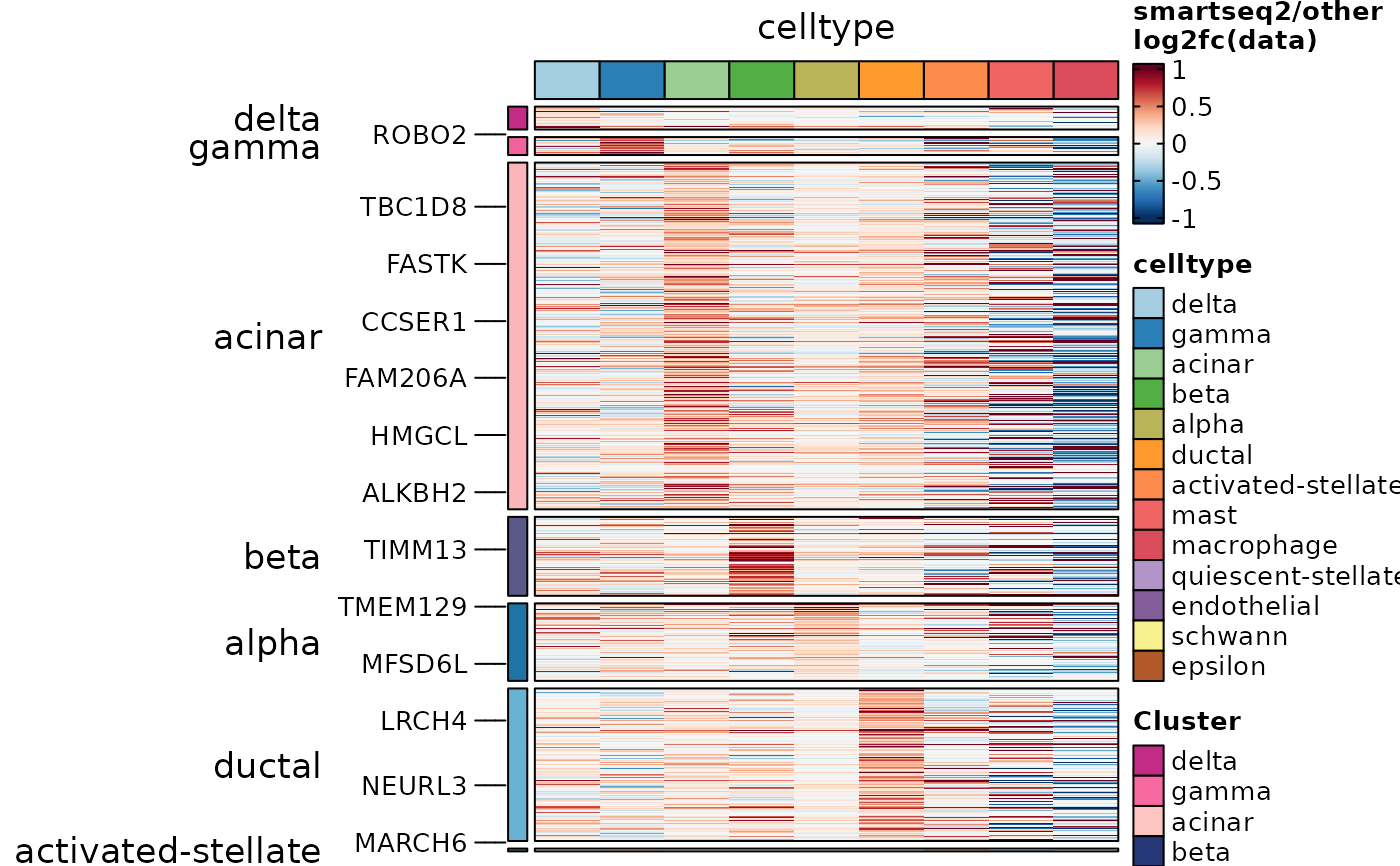 ht7$plot
ht7$plot