Perform the enrichment analysis (over-representation) on the genes
Source:R/RunEnrichment.R
RunEnrichment.RdPerform the enrichment analysis (over-representation) on the genes
Usage
RunEnrichment(
srt = NULL,
group.by = NULL,
test.use = "wilcox",
DE_threshold = "avg_log2FC > 0 & p_val_adj < 0.05",
geneID = NULL,
geneID_groups = NULL,
geneID_exclude = NULL,
IDtype = "symbol",
result_IDtype = "symbol",
species = "Homo_sapiens",
db = "GO_BP",
db_update = FALSE,
db_version = "latest",
db_combine = FALSE,
convert_species = TRUE,
Ensembl_version = NULL,
mirror = NULL,
TERM2GENE = NULL,
TERM2NAME = NULL,
minGSSize = 10,
maxGSSize = 500,
unlimited_db = c("Chromosome", "GeneType", "TF", "Enzyme", "CSPA"),
GO_simplify = FALSE,
GO_simplify_cutoff = "p.adjust < 0.05",
simplify_method = "Wang",
simplify_similarityCutoff = 0.7,
cores = 1,
verbose = TRUE
)Arguments
- srt
A Seurat object containing the results of differential expression analysis (RunDEtest). If specified, the genes and groups will be extracted from the Seurat object automatically. If not specified, the
geneIDandgeneID_groupsarguments must be provided.- group.by
Name of one or more meta.data columns to group (color) cells by.
- test.use
A character vector specifying the test to be used in differential expression analysis. This argument is only used if
srtis specified.- DE_threshold
A character vector specifying the filter condition for differential expression analysis. This argument is only used if
srtis specified.- geneID
A character vector specifying the gene IDs.
- geneID_groups
A factor vector specifying the group labels for each gene.
- geneID_exclude
A character vector specifying the gene IDs to be excluded from the analysis.
- IDtype
A character vector specifying the type of gene IDs in the
srtobject orgeneIDargument. This argument is used to convert the gene IDs to a different type ifIDtypeis different fromresult_IDtype.- result_IDtype
A character vector specifying the desired type of gene ID to be used in the output. This argument is used to convert the gene IDs from
IDtypetoresult_IDtype.- species
A character vector specifying the species for which the gene annotation databases should be prepared. Can be
"Homo_sapiens"or"Mus_musculus".- db
A character vector specifying the annotation sources to be included in the gene annotation databases. Can be one or more of
"GO", "GO_BP", "GO_CC", "GO_MF", "KEGG", "WikiPathway", "Reactome", "CORUM", "MP", "DO", "HPO", "PFAM", "CSPA", "Surfaceome", "SPRomeDB", "VerSeDa", "TFLink", "hTFtarget", "TRRUST", "JASPAR", "ENCODE", "MSigDB", "CellTalk", "CellChat", "Chromosome", "GeneType", "Enzyme", "TF".- db_update
Whether the gene annotation databases should be forcefully updated. If set to FALSE, the function will attempt to load the cached databases instead. Default is
FALSE.- db_version
A character vector specifying the version of the gene annotation databases to be retrieved. Default is
"latest".- db_combine
Whether to combine multiple databases into one. If
TRUE, all database specified bydbwill be combined as one named "Combined".- convert_species
Whether to use a species-converted database when the annotation is missing for the specified species. Default is
TRUE.- Ensembl_version
An integer specifying the Ensembl version. Default is
NULL. IfNULL, the latest version will be used.- mirror
Specify an Ensembl mirror to connect to. The valid options here are
"www","uswest","useast","asia".- TERM2GENE
A data frame specifying the gene-term mapping for a custom database. The first column should contain the term IDs, and the second column should contain the gene IDs.
- TERM2NAME
A data frame specifying the term-name mapping for a custom database. The first column should contain the term IDs, and the second column should contain the corresponding term names.
- minGSSize
The minimum size of a gene set to be considered in the enrichment analysis.
- maxGSSize
The maximum size of a gene set to be considered in the enrichment analysis.
- unlimited_db
A character vector specifying the names of databases that do not have size restrictions.
- GO_simplify
Whether to simplify the GO terms. If
TRUE, additional results with simplified GO terms will be returned.- GO_simplify_cutoff
A character vector specifying the filter condition for simplification of GO terms. This argument is only used if
GO_simplifyisTRUE.- simplify_method
A character vector specifying the method to be used for simplification of GO terms. This argument is only used if
GO_simplifyisTRUE.- simplify_similarityCutoff
The similarity cutoff for simplification of GO terms. This argument is only used if
GO_simplifyisTRUE.- cores
The number of cores to use for parallelization with foreach::foreach. Default is
1.- verbose
Whether to print the message. Default is
TRUE.
Value
If input is a Seurat object, returns the modified Seurat object with the enrichment result stored in the tools slot.
If input is a geneID vector with or without geneID_groups, return the enrichment result directly.
Enrichment result is a list with the following component:
enrichment: A data.frame containing all enrichment results.results: A list ofenrichResultobjects from the DOSE package.geneMap: A data.frame containing the ID mapping table for input gene IDs.input: A data.frame containing the input gene IDs and gene ID groups.DE_threshold: A specific threshold for differential expression analysis (only returned if input is a Seurat object).
Examples
data(pancreas_sub)
pancreas_sub <- standard_scop(pancreas_sub)
#> ℹ [2026-02-11 03:58:58] Start standard scop workflow...
#> ℹ [2026-02-11 03:58:58] Checking a list of <Seurat>...
#> ! [2026-02-11 03:58:58] Data 1/1 of the `srt_list` is "unknown"
#> ℹ [2026-02-11 03:58:58] Perform `NormalizeData()` with `normalization.method = 'LogNormalize'` on the data 1/1 of the `srt_list`...
#> ℹ [2026-02-11 03:59:01] Perform `Seurat::FindVariableFeatures()` on the data 1/1 of the `srt_list`...
#> ℹ [2026-02-11 03:59:01] Use the separate HVF from srt_list
#> ℹ [2026-02-11 03:59:01] Number of available HVF: 2000
#> ℹ [2026-02-11 03:59:01] Finished check
#> ℹ [2026-02-11 03:59:02] Perform `Seurat::ScaleData()`
#> ℹ [2026-02-11 03:59:02] Perform pca linear dimension reduction
#> ℹ [2026-02-11 03:59:03] Perform `Seurat::FindClusters()` with `cluster_algorithm = 'louvain'` and `cluster_resolution = 0.6`
#> ℹ [2026-02-11 03:59:03] Reorder clusters...
#> ℹ [2026-02-11 03:59:03] Perform umap nonlinear dimension reduction
#> ℹ [2026-02-11 03:59:03] Non-linear dimensionality reduction (umap) using (Standardpca) dims (1-50) as input
#> ℹ [2026-02-11 03:59:08] Non-linear dimensionality reduction (umap) using (Standardpca) dims (1-50) as input
#> ✔ [2026-02-11 03:59:12] Run scop standard workflow completed
pancreas_sub <- RunDEtest(
pancreas_sub,
group.by = "CellType"
)
#> ℹ [2026-02-11 03:59:12] Data type is log-normalized
#> ℹ [2026-02-11 03:59:12] Start differential expression test
#> ℹ [2026-02-11 03:59:12] Find all markers(wilcox) among [1] 5 groups...
#> ℹ [2026-02-11 03:59:12] Using 1 core
#> ⠙ [2026-02-11 03:59:12] Running for Ductal [1/5] ■■■■■■■ …
#> ✔ [2026-02-11 03:59:12] Completed 5 tasks in 720ms
#>
#> ℹ [2026-02-11 03:59:12] Building results
#> ✔ [2026-02-11 03:59:13] Differential expression test completed
pancreas_sub <- RunEnrichment(
pancreas_sub,
group.by = "CellType",
DE_threshold = "p_val_adj < 0.05",
db = "GO_BP",
species = "Mus_musculus"
)
#> ℹ [2026-02-11 03:59:13] Start Enrichment analysis
#> ℹ [2026-02-11 03:59:13] Species: "Mus_musculus"
#> ℹ [2026-02-11 03:59:13] Loading cached: GO_BP version: 3.22.0 nterm:15169 created: 2026-02-11 03:27:59
#> ℹ [2026-02-11 03:59:14] Permform enrichment...
#> ℹ [2026-02-11 03:59:14] Using 1 core
#> ⠙ [2026-02-11 03:59:14] Running for 1 [1/5] ■■■■■■■ 2…
#> ⠹ [2026-02-11 03:59:14] Running for 2 [2/5] ■■■■■■■■■■■■■ 4…
#> ⠸ [2026-02-11 03:59:14] Running for 3 [3/5] ■■■■■■■■■■■■■■■■■■■ 6…
#> ⠼ [2026-02-11 03:59:14] Running for 4 [4/5] ■■■■■■■■■■■■■■■■■■■■■■■■■ 8…
#> ✔ [2026-02-11 03:59:14] Completed 5 tasks in 1m 20s
#>
#> ℹ [2026-02-11 03:59:14] Building results
#> ✔ [2026-02-11 04:00:35] Enrichment analysis done
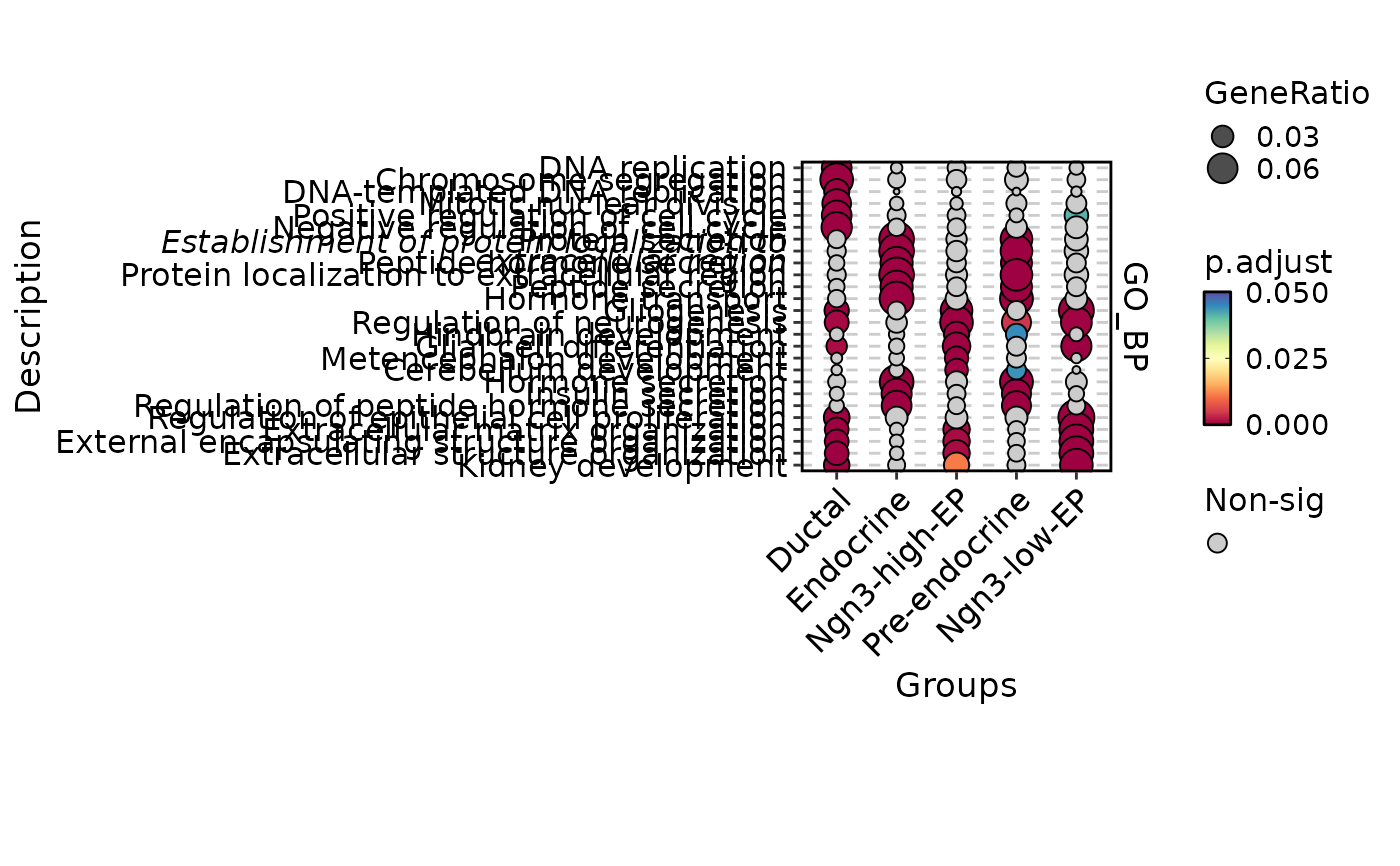
EnrichmentPlot(
pancreas_sub,
db = "GO_BP",
group.by = "CellType",
plot_type = "comparison"
)
 if (FALSE) { # \dontrun{
pancreas_sub <- RunEnrichment(
pancreas_sub,
group.by = "CellType",
DE_threshold = "p_val_adj < 0.05",
db = c("MSigDB", "MSigDB_MH"),
species = "Mus_musculus"
)
EnrichmentPlot(
pancreas_sub,
db = "MSigDB",
group.by = "CellType",
plot_type = "comparison"
)
EnrichmentPlot(
pancreas_sub,
db = "MSigDB_MH",
group.by = "CellType",
plot_type = "comparison"
)
# Remove redundant GO terms
pancreas_sub <- RunEnrichment(
pancreas_sub,
group.by = "CellType",
db = "GO_BP",
GO_simplify = TRUE,
species = "Mus_musculus"
)
EnrichmentPlot(
pancreas_sub,
db = "GO_BP_sim",
group.by = "CellType",
plot_type = "comparison"
)
# Or use "geneID" and "geneID_groups" as input to run enrichment
de_df <- dplyr::filter(
pancreas_sub@tools$DEtest_CellType$AllMarkers_wilcox,
p_val_adj < 0.05
)
enrich_out <- RunEnrichment(
geneID = de_df[["gene"]],
geneID_groups = de_df[["group1"]],
db = "GO_BP",
species = "Mus_musculus"
)
EnrichmentPlot(
res = enrich_out,
db = "GO_BP",
plot_type = "comparison"
)
# Use a combined database
pancreas_sub <- RunEnrichment(
pancreas_sub,
group.by = "CellType",
db = c(
"KEGG", "WikiPathway", "Reactome", "PFAM", "MP"
),
db_combine = TRUE,
species = "Mus_musculus"
)
EnrichmentPlot(
pancreas_sub,
db = "Combined",
group.by = "CellType",
plot_type = "comparison"
)
} # }
if (FALSE) { # \dontrun{
pancreas_sub <- RunEnrichment(
pancreas_sub,
group.by = "CellType",
DE_threshold = "p_val_adj < 0.05",
db = c("MSigDB", "MSigDB_MH"),
species = "Mus_musculus"
)
EnrichmentPlot(
pancreas_sub,
db = "MSigDB",
group.by = "CellType",
plot_type = "comparison"
)
EnrichmentPlot(
pancreas_sub,
db = "MSigDB_MH",
group.by = "CellType",
plot_type = "comparison"
)
# Remove redundant GO terms
pancreas_sub <- RunEnrichment(
pancreas_sub,
group.by = "CellType",
db = "GO_BP",
GO_simplify = TRUE,
species = "Mus_musculus"
)
EnrichmentPlot(
pancreas_sub,
db = "GO_BP_sim",
group.by = "CellType",
plot_type = "comparison"
)
# Or use "geneID" and "geneID_groups" as input to run enrichment
de_df <- dplyr::filter(
pancreas_sub@tools$DEtest_CellType$AllMarkers_wilcox,
p_val_adj < 0.05
)
enrich_out <- RunEnrichment(
geneID = de_df[["gene"]],
geneID_groups = de_df[["group1"]],
db = "GO_BP",
species = "Mus_musculus"
)
EnrichmentPlot(
res = enrich_out,
db = "GO_BP",
plot_type = "comparison"
)
# Use a combined database
pancreas_sub <- RunEnrichment(
pancreas_sub,
group.by = "CellType",
db = c(
"KEGG", "WikiPathway", "Reactome", "PFAM", "MP"
),
db_combine = TRUE,
species = "Mus_musculus"
)
EnrichmentPlot(
pancreas_sub,
db = "Combined",
group.by = "CellType",
plot_type = "comparison"
)
} # }