This function performs KNN prediction to annotate cell types based on reference scRNA-seq or bulk RNA-seq data.
Usage
RunKNNPredict(
srt_query,
srt_ref = NULL,
bulk_ref = NULL,
query_group = NULL,
ref_group = NULL,
query_assay = NULL,
ref_assay = NULL,
query_reduction = NULL,
ref_reduction = NULL,
query_dims = 1:30,
ref_dims = 1:30,
query_collapsing = !is.null(query_group),
ref_collapsing = TRUE,
return_full_distance_matrix = FALSE,
features = NULL,
features_type = c("HVF", "DE"),
feature_source = "both",
nfeatures = 2000,
DEtest_param = list(max.cells.per.ident = 200, test.use = "wilcox"),
DE_threshold = "p_val_adj < 0.05",
nn_method = NULL,
distance_metric = "cosine",
k = 30,
filter_lowfreq = 0,
prefix = "KNNPredict"
)Arguments
- srt_query
An object of class Seurat to be annotated with cell types.
- srt_ref
An object of class Seurat storing the reference cells.
- bulk_ref
A cell atlas matrix, where cell types are represented by columns and genes are represented by rows, for example, scop::ref_scHCL. Either
srt_reforbulk_refmust be provided.- query_group
A character vector specifying the column name in the
srt_querymetadata that represents the cell grouping.- ref_group
A character vector specifying the column name in the
srt_refmetadata that represents the cell grouping.- query_assay
A character vector specifying the assay to be used for the query data. Default is the default assay of the
srt_queryobject.- ref_assay
A character vector specifying the assay to be used for the reference data. Default is the default assay of the
srt_refobject.- query_reduction
A character vector specifying the dimensionality reduction method used for the query data. If NULL, the function will use the default reduction method specified in the
srt_queryobject.- ref_reduction
A character vector specifying the dimensionality reduction method used for the reference data. If NULL, the function will use the default reduction method specified in the
srt_refobject.- query_dims
A numeric vector specifying the dimensions to be used for the query data. Default is the first
30dimensions.- ref_dims
A numeric vector specifying the dimensions to be used for the reference data. Default is the first
30dimensions.- query_collapsing
A boolean value indicating whether the query data should be collapsed to group-level average expression values. If TRUE, the function will calculate the average expression values for each group in the query data and the annotation will be performed separately for each group. Otherwise it will use the raw expression values for each cell.
- ref_collapsing
A boolean value indicating whether the reference data should be collapsed to group-level average expression values. If TRUE, the function will calculate the average expression values for each group in the reference data and the annotation will be performed separately for each group. Otherwise it will use the raw expression values for each cell.
- return_full_distance_matrix
A boolean value indicating whether the full distance matrix should be returned. If TRUE, the function will return the distance matrix used for the KNN prediction, otherwise it will only return the annotated cell types.
- features
A character vector specifying the features to be used for the KNN prediction. If
NULL, all the features in the query and reference data will be used.- features_type
A character vector specifying the type of features to be used for the KNN prediction. Must be one of "HVF" (highly variable features) or "DE" (differentially expressed features). Default is
"HVF".- feature_source
The source of the features to be used. Must be one of "both", "query", or "ref". Default is
"both".- nfeatures
An integer specifying the maximum number of features to be used for the KNN prediction. Default is
2000.- DEtest_param
A list of parameters to be passed to the differential expression test function if
features_typeis set to "DE". Default islist(max.cells.per.ident = 200, test.use = "wilcox").- DE_threshold
Threshold used to filter the DE features. If using "roc" test,
DE_thresholdshould be needs to be reassigned. e.g. "power > 0.5". Default is"p_val < 0.05".- nn_method
A character string specifying the nearest neighbor search method to use. Options are "raw", "annoy", and "rann". If "raw" is selected, the function will use the brute-force method to find the nearest neighbors. If "annoy" is selected, the function will use the Annoy library for approximate nearest neighbor search. If "rann" is selected, the function will use the RANN library for approximate nearest neighbor search. If not provided, the function will choose the search method based on the size of the query and reference datasets.
- distance_metric
A character vector specifying the distance metric to be used for calculating similarity between cells. Must be one of "cosine", "euclidean", "manhattan", or "hamming". Default is
"cosine".- k
A number of nearest neighbors to be considered for the KNN prediction. Default is
30.- filter_lowfreq
An integer specifying the threshold for filtering low-frequency cell types from the predicted results. Cell types with a frequency lower than
filter_lowfreqwill be labelled as "unreliable". Default is0, which means no filtering will be performed.- prefix
A character vector specifying the prefix to be added to the resulting annotations. Default is
"KNNPredict".
Examples
# Annotate cells using bulk RNA-seq data
data(pancreas_sub)
data(ref_scMCA)
pancreas_sub <- standard_scop(pancreas_sub)
#> ℹ [2026-02-11 04:02:50] Start standard scop workflow...
#> ℹ [2026-02-11 04:02:51] Checking a list of <Seurat>...
#> ! [2026-02-11 04:02:51] Data 1/1 of the `srt_list` is "unknown"
#> ℹ [2026-02-11 04:02:51] Perform `NormalizeData()` with `normalization.method = 'LogNormalize'` on the data 1/1 of the `srt_list`...
#> ℹ [2026-02-11 04:02:53] Perform `Seurat::FindVariableFeatures()` on the data 1/1 of the `srt_list`...
#> ℹ [2026-02-11 04:02:53] Use the separate HVF from srt_list
#> ℹ [2026-02-11 04:02:53] Number of available HVF: 2000
#> ℹ [2026-02-11 04:02:54] Finished check
#> ℹ [2026-02-11 04:02:54] Perform `Seurat::ScaleData()`
#> ℹ [2026-02-11 04:02:54] Perform pca linear dimension reduction
#> ℹ [2026-02-11 04:02:55] Perform `Seurat::FindClusters()` with `cluster_algorithm = 'louvain'` and `cluster_resolution = 0.6`
#> ℹ [2026-02-11 04:02:55] Reorder clusters...
#> ℹ [2026-02-11 04:02:55] Perform umap nonlinear dimension reduction
#> ℹ [2026-02-11 04:02:55] Non-linear dimensionality reduction (umap) using (Standardpca) dims (1-50) as input
#> ℹ [2026-02-11 04:03:00] Non-linear dimensionality reduction (umap) using (Standardpca) dims (1-50) as input
#> ✔ [2026-02-11 04:03:04] Run scop standard workflow completed
# Set the number of threads for RcppParallel
# details see: ?RcppParallel::setThreadOptions
# if (requireNamespace("RcppParallel", quietly = TRUE)) {
# RcppParallel::setThreadOptions()
# }
pancreas_sub <- RunKNNPredict(
srt_query = pancreas_sub,
bulk_ref = ref_scMCA
)
#> ℹ [2026-02-11 04:03:04] Use [1] 549 features to calculate distance.
#> ℹ [2026-02-11 04:03:04] Detected query data type: "log_normalized_counts"
#> ℹ [2026-02-11 04:03:04] Detected reference data type: "log_normalized_counts"
#> ℹ [2026-02-11 04:03:04] Calculate similarity...
#> ℹ [2026-02-11 04:03:04] Use raw method to find neighbors
#> ℹ [2026-02-11 04:03:05] Predict cell type...
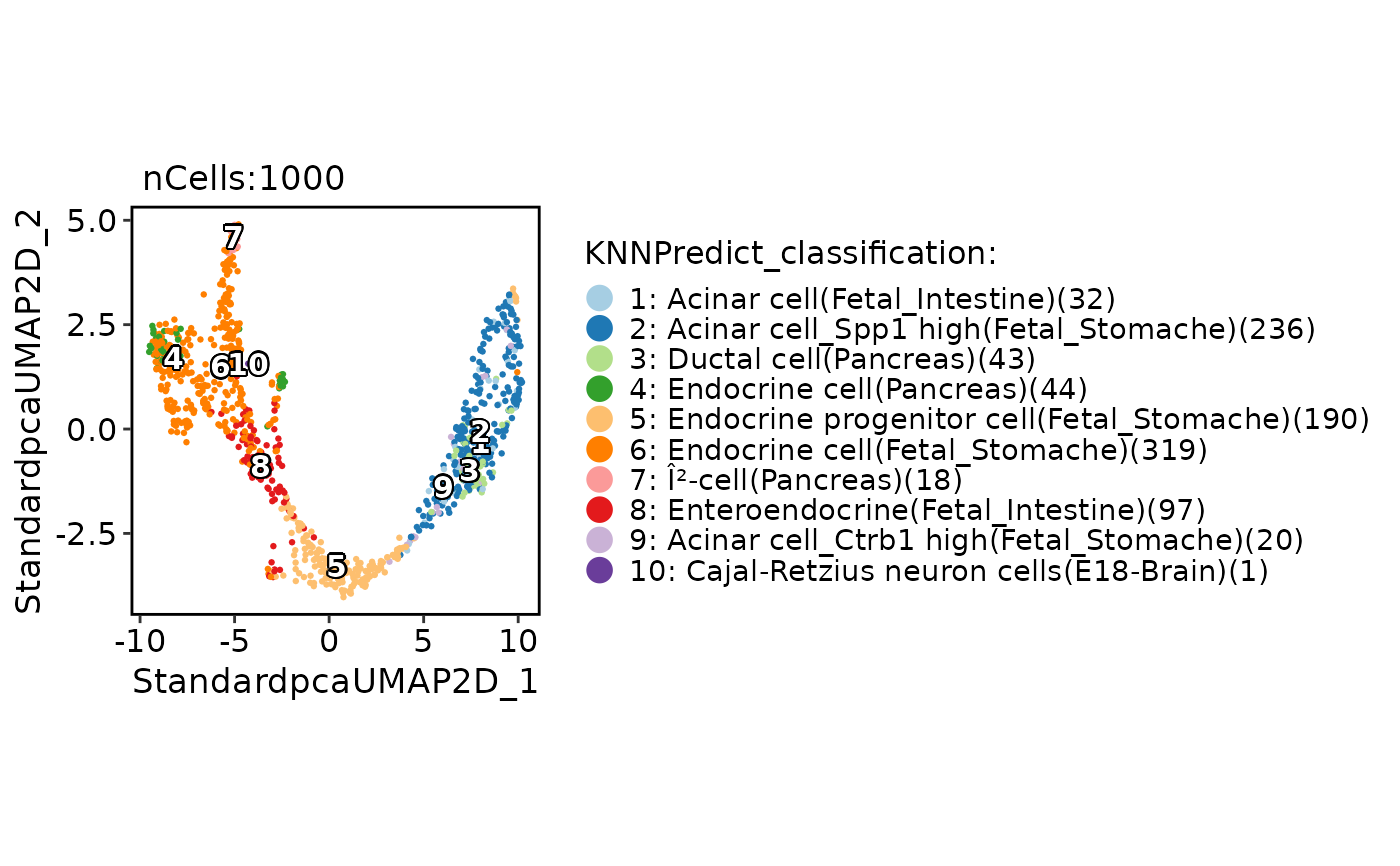
CellDimPlot(
pancreas_sub,
group.by = "KNNPredict_classification",
label = TRUE
)
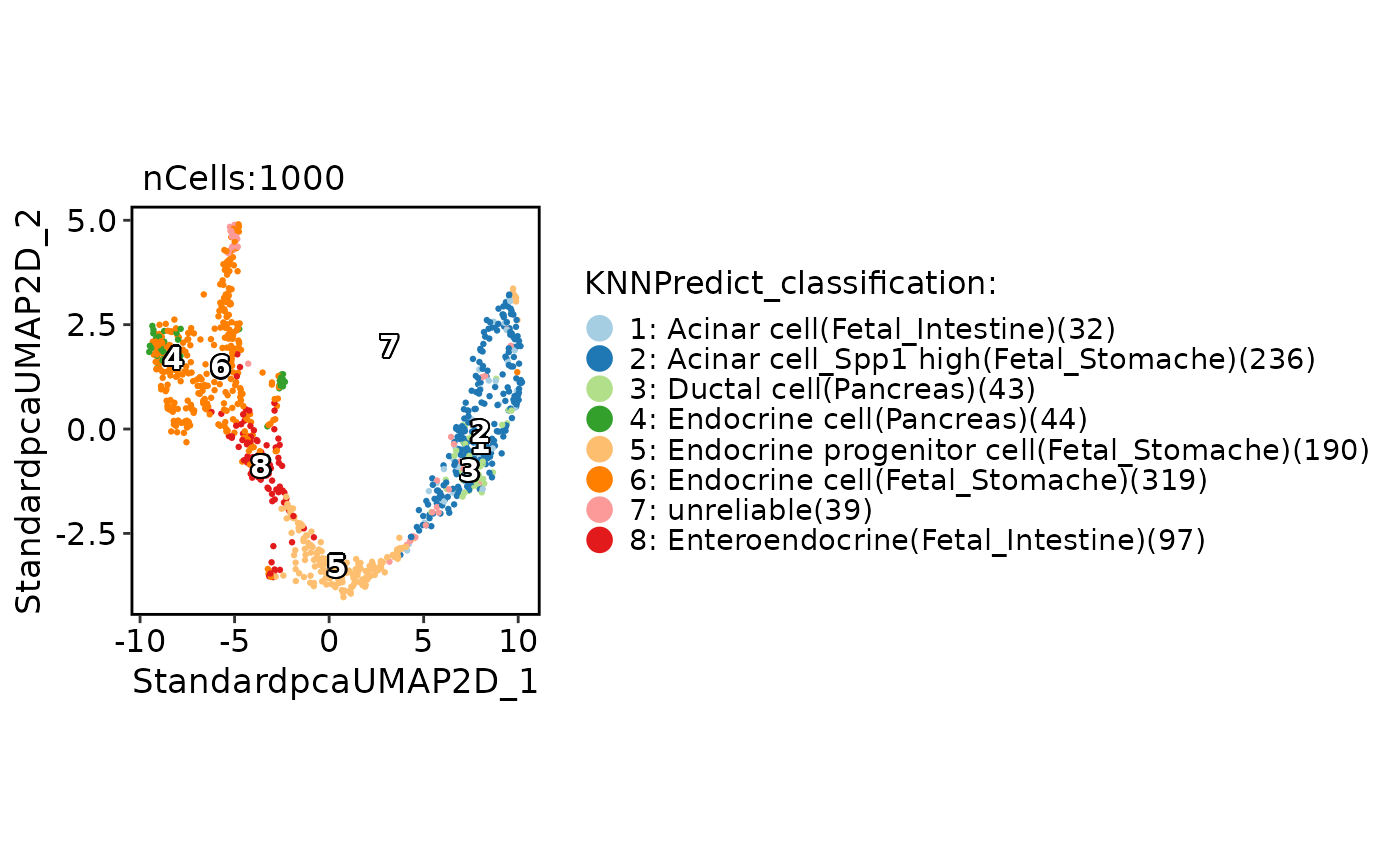
 # Removal of low credible cell types from the predicted results
pancreas_sub <- RunKNNPredict(
srt_query = pancreas_sub,
bulk_ref = ref_scMCA,
filter_lowfreq = 30
)
#> ℹ [2026-02-11 04:03:05] Use [1] 549 features to calculate distance.
#> ℹ [2026-02-11 04:03:05] Detected query data type: "log_normalized_counts"
#> ℹ [2026-02-11 04:03:05] Detected reference data type: "log_normalized_counts"
#> ℹ [2026-02-11 04:03:05] Calculate similarity...
#> ℹ [2026-02-11 04:03:05] Use raw method to find neighbors
#> ℹ [2026-02-11 04:03:06] Predict cell type...
CellDimPlot(
pancreas_sub,
group.by = "KNNPredict_classification",
label = TRUE
)
# Removal of low credible cell types from the predicted results
pancreas_sub <- RunKNNPredict(
srt_query = pancreas_sub,
bulk_ref = ref_scMCA,
filter_lowfreq = 30
)
#> ℹ [2026-02-11 04:03:05] Use [1] 549 features to calculate distance.
#> ℹ [2026-02-11 04:03:05] Detected query data type: "log_normalized_counts"
#> ℹ [2026-02-11 04:03:05] Detected reference data type: "log_normalized_counts"
#> ℹ [2026-02-11 04:03:05] Calculate similarity...
#> ℹ [2026-02-11 04:03:05] Use raw method to find neighbors
#> ℹ [2026-02-11 04:03:06] Predict cell type...
CellDimPlot(
pancreas_sub,
group.by = "KNNPredict_classification",
label = TRUE
)
 # Annotate clusters using bulk RNA-seq data
pancreas_sub <- RunKNNPredict(
srt_query = pancreas_sub,
query_group = "SubCellType",
bulk_ref = ref_scMCA
)
#> ℹ [2026-02-11 04:03:06] Use [1] 549 features to calculate distance.
#> ℹ [2026-02-11 04:03:06] Detected query data type: "log_normalized_counts"
#> ℹ [2026-02-11 04:03:06] Detected reference data type: "log_normalized_counts"
#> ℹ [2026-02-11 04:03:06] Calculate similarity...
#> ℹ [2026-02-11 04:03:06] Use raw method to find neighbors
#> ℹ [2026-02-11 04:03:06] Predict cell type...
CellDimPlot(
pancreas_sub,
group.by = "KNNPredict_classification",
label = TRUE
)
# Annotate clusters using bulk RNA-seq data
pancreas_sub <- RunKNNPredict(
srt_query = pancreas_sub,
query_group = "SubCellType",
bulk_ref = ref_scMCA
)
#> ℹ [2026-02-11 04:03:06] Use [1] 549 features to calculate distance.
#> ℹ [2026-02-11 04:03:06] Detected query data type: "log_normalized_counts"
#> ℹ [2026-02-11 04:03:06] Detected reference data type: "log_normalized_counts"
#> ℹ [2026-02-11 04:03:06] Calculate similarity...
#> ℹ [2026-02-11 04:03:06] Use raw method to find neighbors
#> ℹ [2026-02-11 04:03:06] Predict cell type...
CellDimPlot(
pancreas_sub,
group.by = "KNNPredict_classification",
label = TRUE
)
 # Annotate using single cell RNA-seq data
data(panc8_sub)
# Simply convert genes from human to mouse and preprocess the data
genenames <- make.unique(
thisutils::capitalize(
rownames(panc8_sub),
force_tolower = TRUE
)
)
names(genenames) <- rownames(panc8_sub)
panc8_sub <- RenameFeatures(
panc8_sub,
newnames = genenames
)
#> ℹ [2026-02-11 04:03:07] Rename features for the assay: RNA
panc8_sub <- CheckDataMerge(
panc8_sub,
batch = "tech"
)[["srt_merge"]]
#> ℹ [2026-02-11 04:03:07] Spliting `srt_merge` into `srt_list` by column "tech"...
#> ℹ [2026-02-11 04:03:08] Checking a list of <Seurat>...
#> ! [2026-02-11 04:03:08] Data 1/5 of the `srt_list` is "unknown"
#> ℹ [2026-02-11 04:03:08] Perform `NormalizeData()` with `normalization.method = 'LogNormalize'` on the data 1/5 of the `srt_list`...
#> ℹ [2026-02-11 04:03:09] Perform `Seurat::FindVariableFeatures()` on the data 1/5 of the `srt_list`...
#> ! [2026-02-11 04:03:10] Data 2/5 of the `srt_list` is "unknown"
#> ℹ [2026-02-11 04:03:10] Perform `NormalizeData()` with `normalization.method = 'LogNormalize'` on the data 2/5 of the `srt_list`...
#> ℹ [2026-02-11 04:03:11] Perform `Seurat::FindVariableFeatures()` on the data 2/5 of the `srt_list`...
#> ! [2026-02-11 04:03:12] Data 3/5 of the `srt_list` is "unknown"
#> ℹ [2026-02-11 04:03:12] Perform `NormalizeData()` with `normalization.method = 'LogNormalize'` on the data 3/5 of the `srt_list`...
#> ℹ [2026-02-11 04:03:13] Perform `Seurat::FindVariableFeatures()` on the data 3/5 of the `srt_list`...
#> ! [2026-02-11 04:03:14] Data 4/5 of the `srt_list` is "unknown"
#> ℹ [2026-02-11 04:03:14] Perform `NormalizeData()` with `normalization.method = 'LogNormalize'` on the data 4/5 of the `srt_list`...
#> ℹ [2026-02-11 04:03:15] Perform `Seurat::FindVariableFeatures()` on the data 4/5 of the `srt_list`...
#> ! [2026-02-11 04:03:16] Data 5/5 of the `srt_list` is "unknown"
#> ℹ [2026-02-11 04:03:16] Perform `NormalizeData()` with `normalization.method = 'LogNormalize'` on the data 5/5 of the `srt_list`...
#> ℹ [2026-02-11 04:03:18] Perform `Seurat::FindVariableFeatures()` on the data 5/5 of the `srt_list`...
#> ℹ [2026-02-11 04:03:18] Use the separate HVF from srt_list
#> ℹ [2026-02-11 04:03:18] Number of available HVF: 2000
#> ℹ [2026-02-11 04:03:19] Finished check
panc8_sub <- SeuratObject::JoinLayers(panc8_sub)
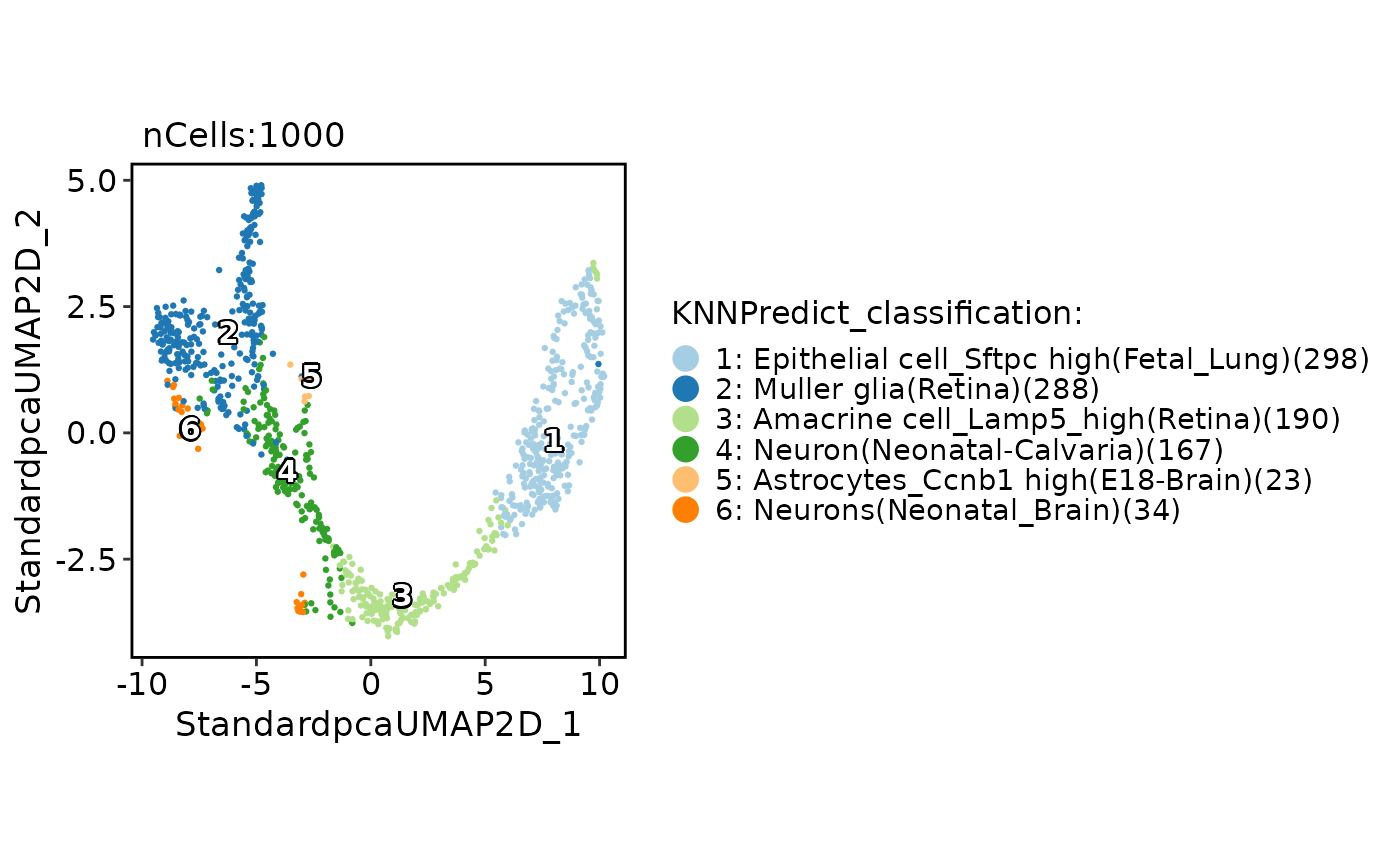
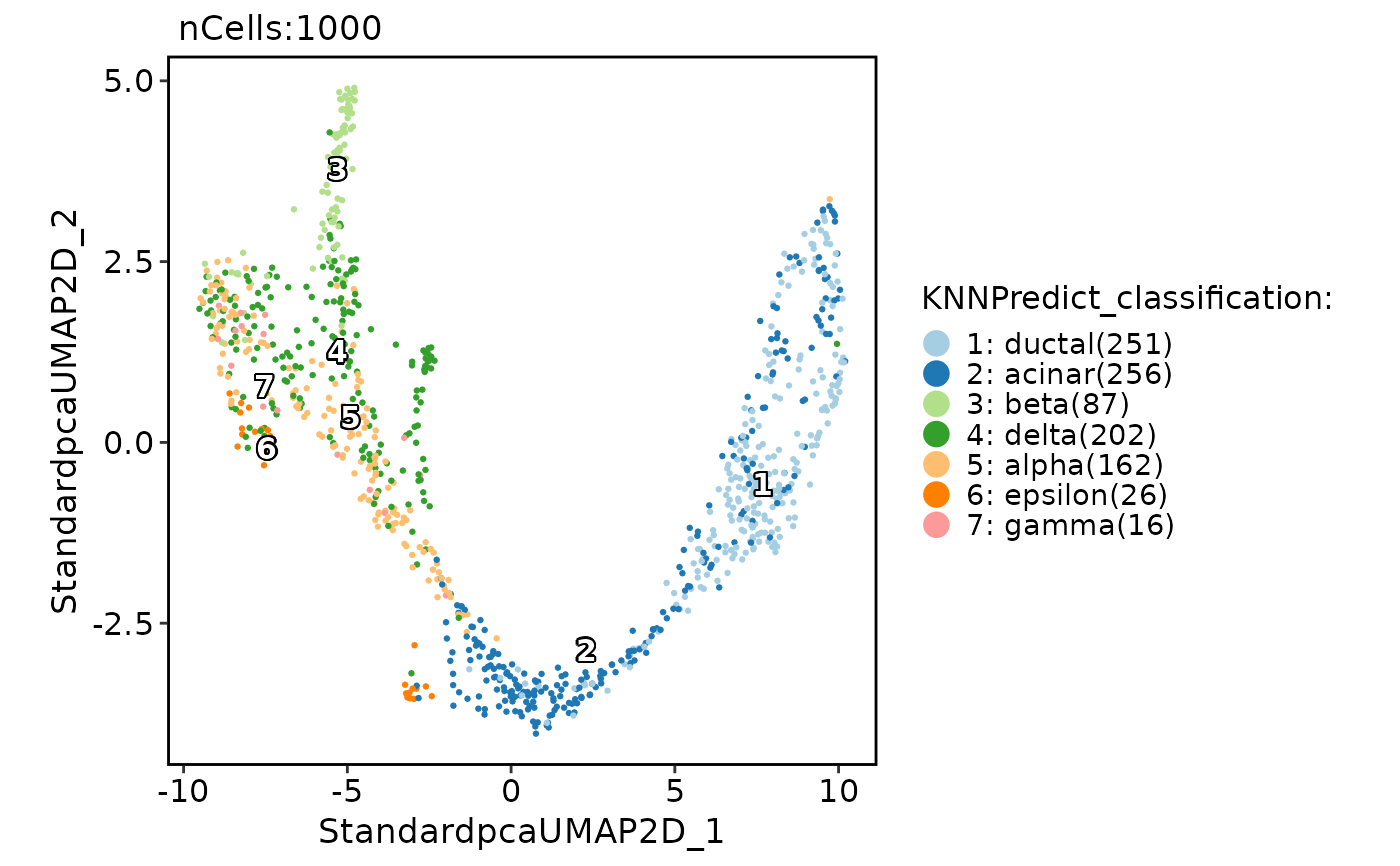
pancreas_sub <- RunKNNPredict(
srt_query = pancreas_sub,
srt_ref = panc8_sub,
ref_group = "celltype"
)
#> ℹ [2026-02-11 04:03:24] Use the HVF to calculate distance metric
#> ℹ [2026-02-11 04:03:24] Use [1] 632 features to calculate distance.
#> ℹ [2026-02-11 04:03:24] Detected query data type: "log_normalized_counts"
#> ℹ [2026-02-11 04:03:24] Detected reference data type: "log_normalized_counts"
#> ℹ [2026-02-11 04:03:24] Calculate similarity...
#> ℹ [2026-02-11 04:03:24] Use raw method to find neighbors
#> ℹ [2026-02-11 04:03:24] Predict cell type...
CellDimPlot(
pancreas_sub,
group.by = "KNNPredict_classification",
label = TRUE
)
# Annotate using single cell RNA-seq data
data(panc8_sub)
# Simply convert genes from human to mouse and preprocess the data
genenames <- make.unique(
thisutils::capitalize(
rownames(panc8_sub),
force_tolower = TRUE
)
)
names(genenames) <- rownames(panc8_sub)
panc8_sub <- RenameFeatures(
panc8_sub,
newnames = genenames
)
#> ℹ [2026-02-11 04:03:07] Rename features for the assay: RNA
panc8_sub <- CheckDataMerge(
panc8_sub,
batch = "tech"
)[["srt_merge"]]
#> ℹ [2026-02-11 04:03:07] Spliting `srt_merge` into `srt_list` by column "tech"...
#> ℹ [2026-02-11 04:03:08] Checking a list of <Seurat>...
#> ! [2026-02-11 04:03:08] Data 1/5 of the `srt_list` is "unknown"
#> ℹ [2026-02-11 04:03:08] Perform `NormalizeData()` with `normalization.method = 'LogNormalize'` on the data 1/5 of the `srt_list`...
#> ℹ [2026-02-11 04:03:09] Perform `Seurat::FindVariableFeatures()` on the data 1/5 of the `srt_list`...
#> ! [2026-02-11 04:03:10] Data 2/5 of the `srt_list` is "unknown"
#> ℹ [2026-02-11 04:03:10] Perform `NormalizeData()` with `normalization.method = 'LogNormalize'` on the data 2/5 of the `srt_list`...
#> ℹ [2026-02-11 04:03:11] Perform `Seurat::FindVariableFeatures()` on the data 2/5 of the `srt_list`...
#> ! [2026-02-11 04:03:12] Data 3/5 of the `srt_list` is "unknown"
#> ℹ [2026-02-11 04:03:12] Perform `NormalizeData()` with `normalization.method = 'LogNormalize'` on the data 3/5 of the `srt_list`...
#> ℹ [2026-02-11 04:03:13] Perform `Seurat::FindVariableFeatures()` on the data 3/5 of the `srt_list`...
#> ! [2026-02-11 04:03:14] Data 4/5 of the `srt_list` is "unknown"
#> ℹ [2026-02-11 04:03:14] Perform `NormalizeData()` with `normalization.method = 'LogNormalize'` on the data 4/5 of the `srt_list`...
#> ℹ [2026-02-11 04:03:15] Perform `Seurat::FindVariableFeatures()` on the data 4/5 of the `srt_list`...
#> ! [2026-02-11 04:03:16] Data 5/5 of the `srt_list` is "unknown"
#> ℹ [2026-02-11 04:03:16] Perform `NormalizeData()` with `normalization.method = 'LogNormalize'` on the data 5/5 of the `srt_list`...
#> ℹ [2026-02-11 04:03:18] Perform `Seurat::FindVariableFeatures()` on the data 5/5 of the `srt_list`...
#> ℹ [2026-02-11 04:03:18] Use the separate HVF from srt_list
#> ℹ [2026-02-11 04:03:18] Number of available HVF: 2000
#> ℹ [2026-02-11 04:03:19] Finished check
panc8_sub <- SeuratObject::JoinLayers(panc8_sub)
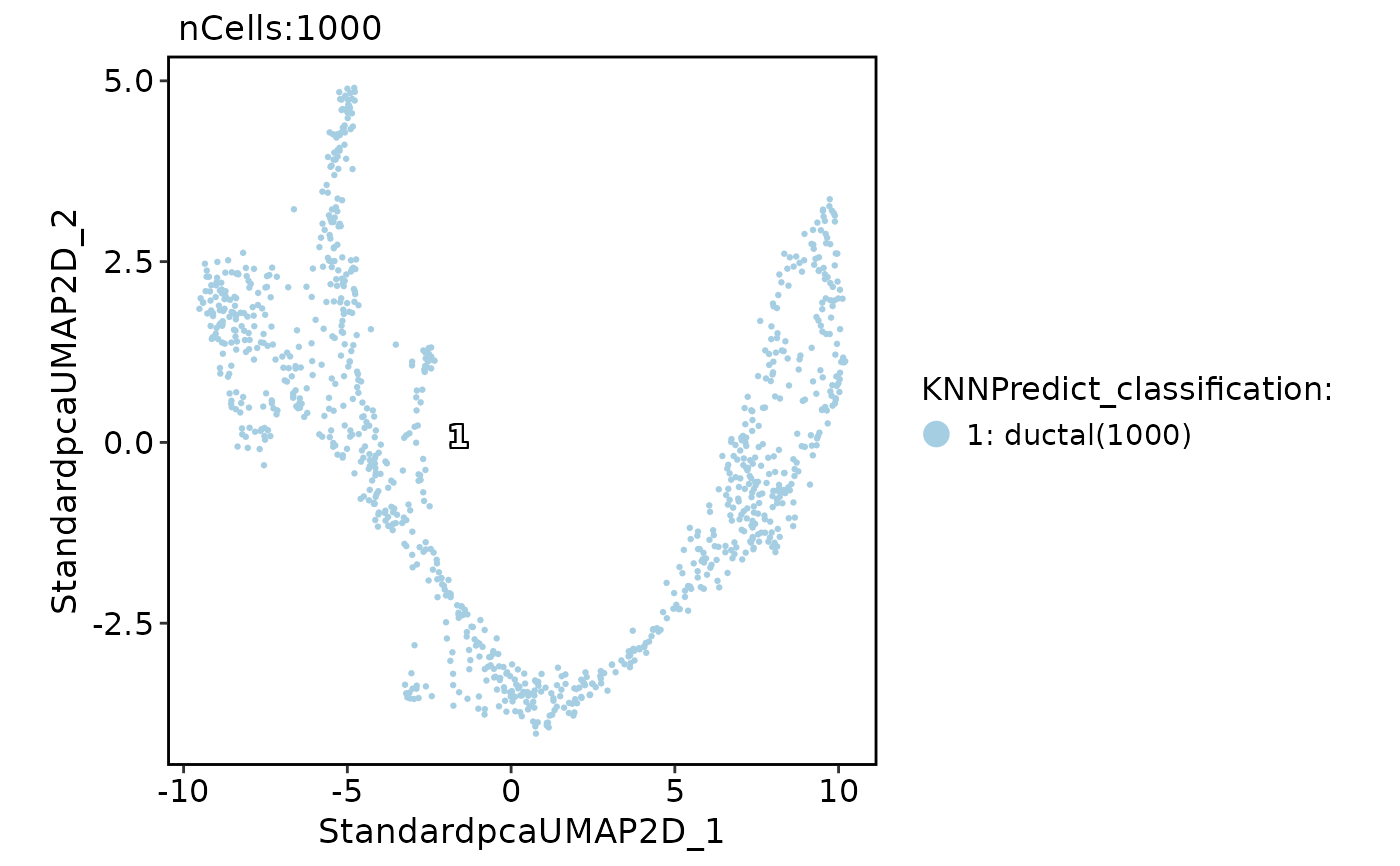
pancreas_sub <- RunKNNPredict(
srt_query = pancreas_sub,
srt_ref = panc8_sub,
ref_group = "celltype"
)
#> ℹ [2026-02-11 04:03:24] Use the HVF to calculate distance metric
#> ℹ [2026-02-11 04:03:24] Use [1] 632 features to calculate distance.
#> ℹ [2026-02-11 04:03:24] Detected query data type: "log_normalized_counts"
#> ℹ [2026-02-11 04:03:24] Detected reference data type: "log_normalized_counts"
#> ℹ [2026-02-11 04:03:24] Calculate similarity...
#> ℹ [2026-02-11 04:03:24] Use raw method to find neighbors
#> ℹ [2026-02-11 04:03:24] Predict cell type...
CellDimPlot(
pancreas_sub,
group.by = "KNNPredict_classification",
label = TRUE
)
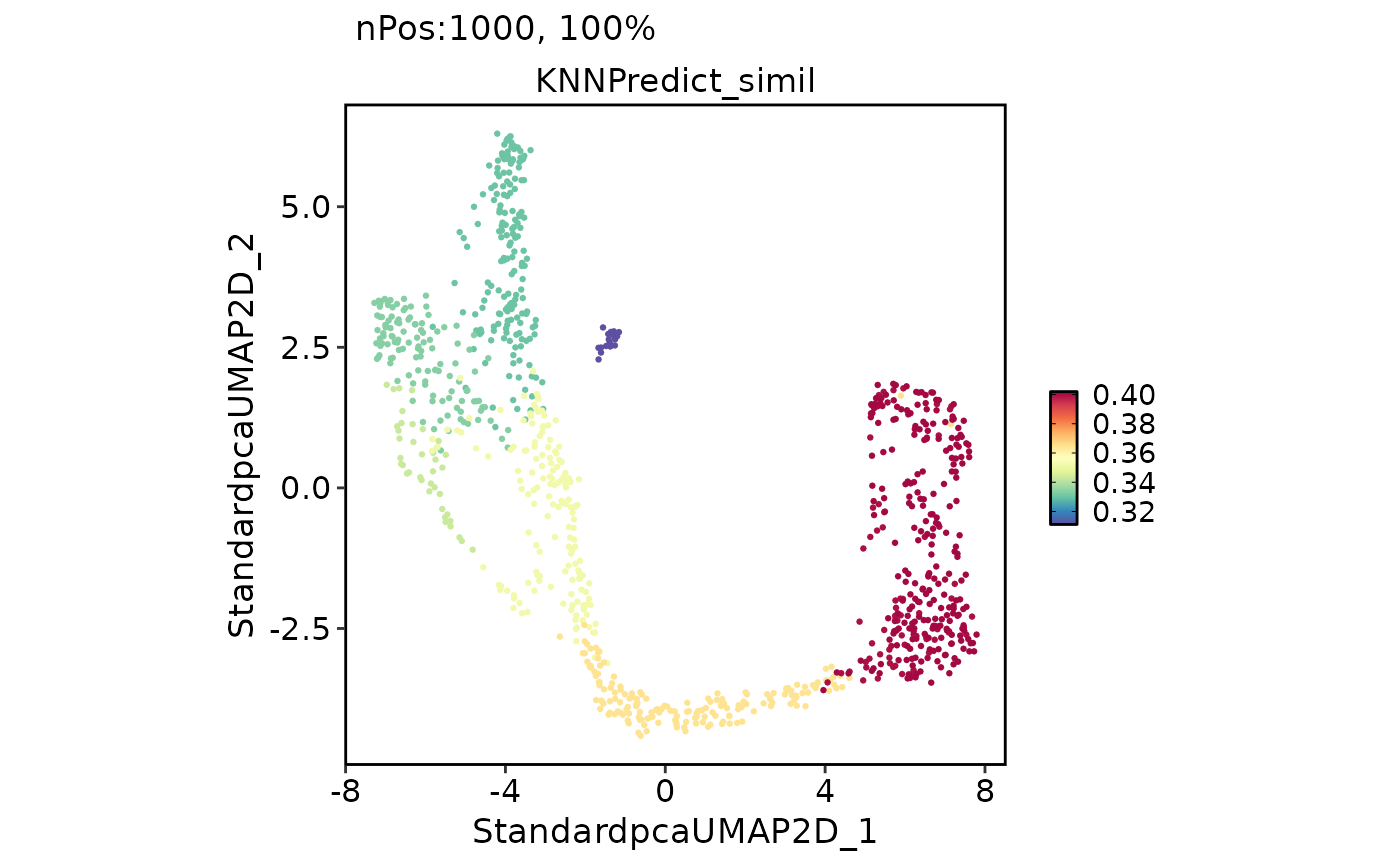
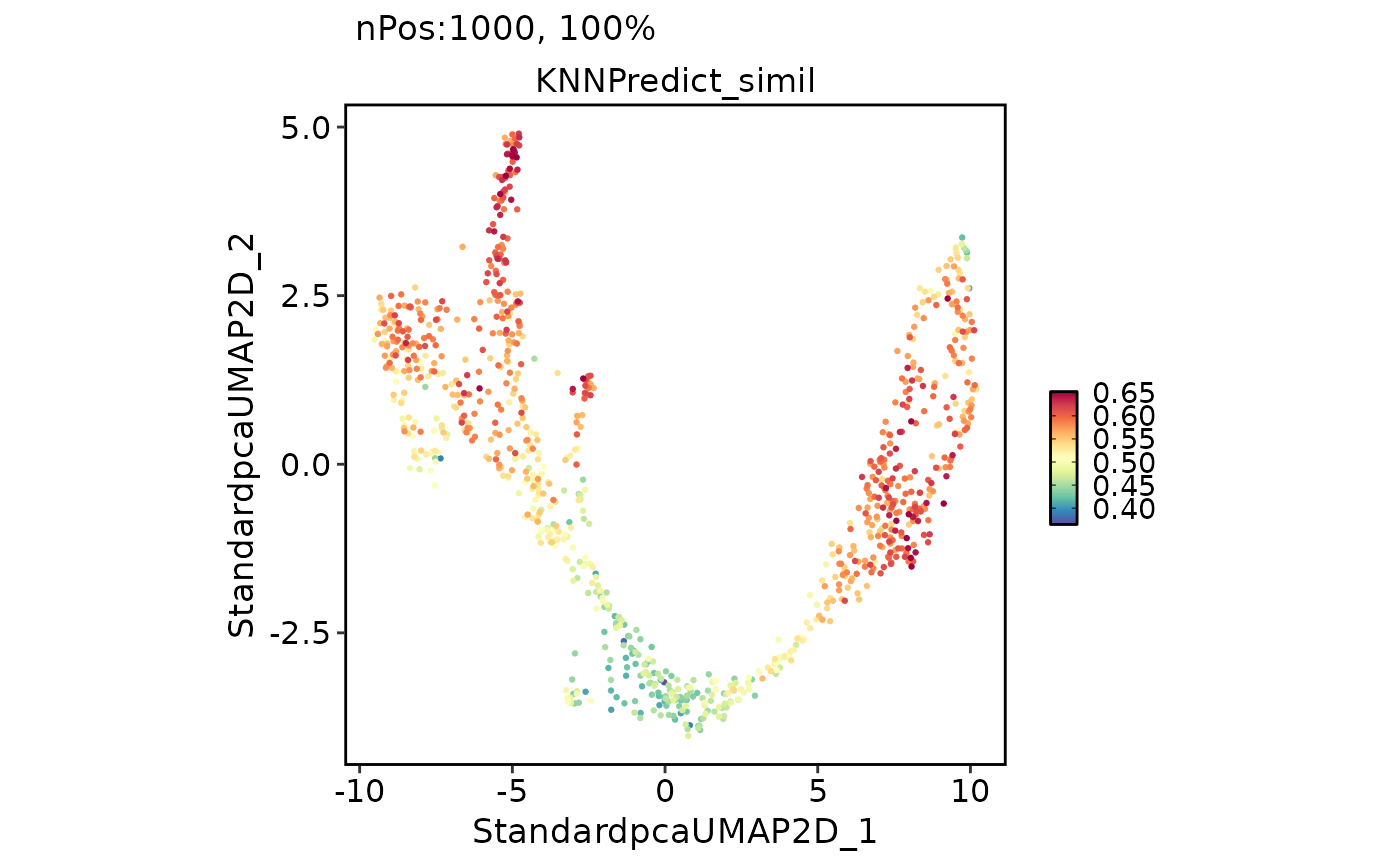
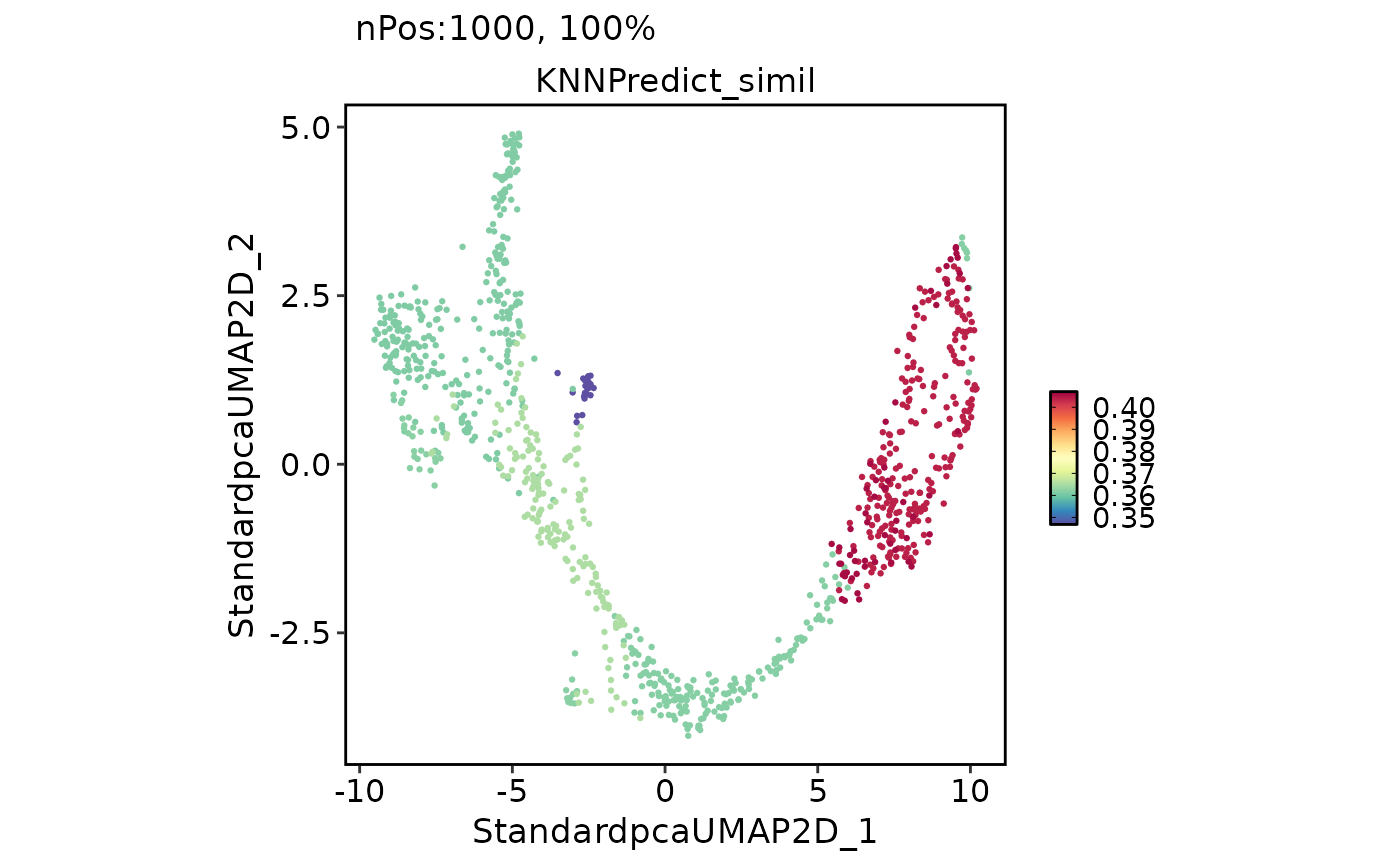
 FeatureDimPlot(
pancreas_sub,
features = "KNNPredict_simil"
)
FeatureDimPlot(
pancreas_sub,
features = "KNNPredict_simil"
)
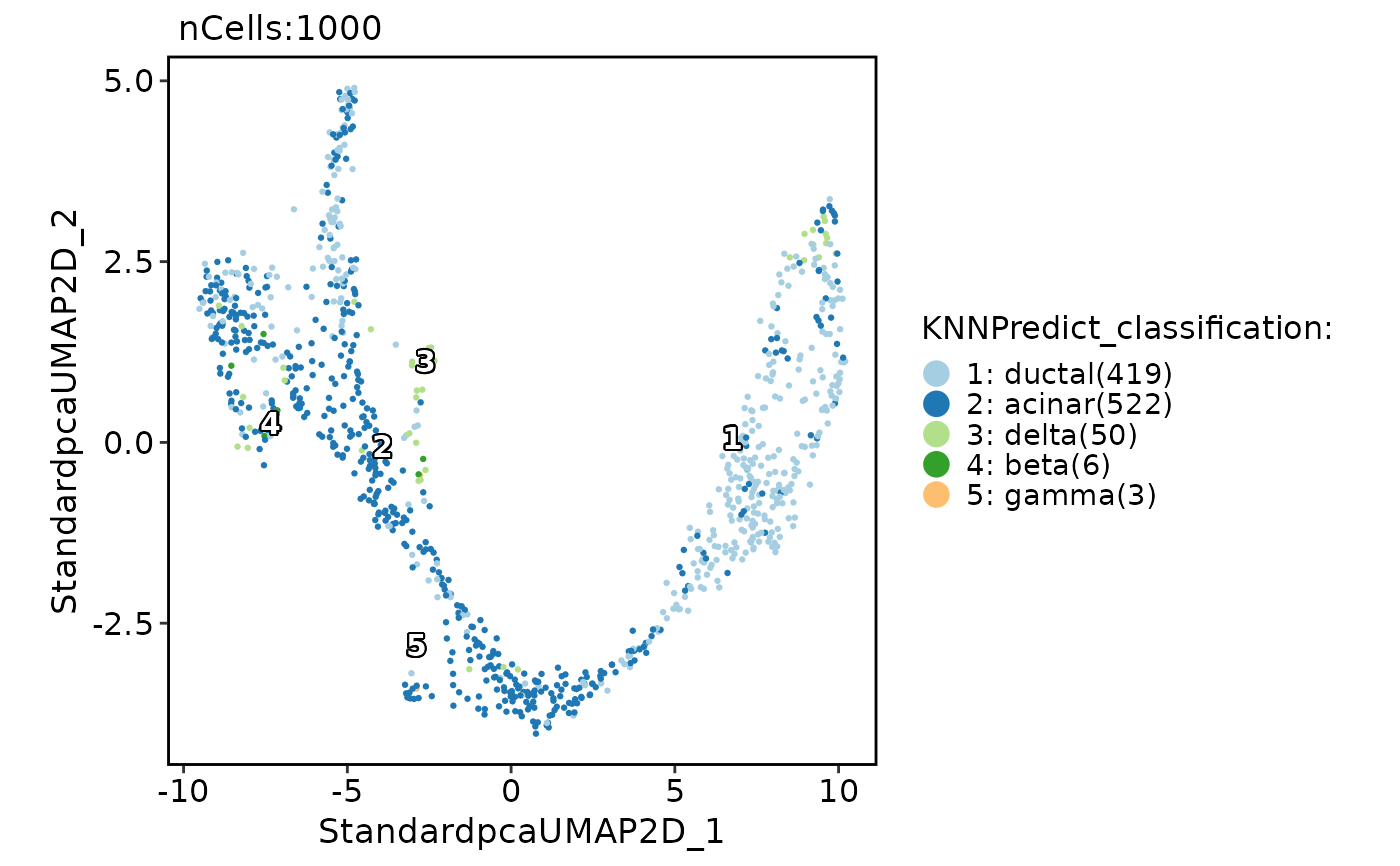
 pancreas_sub <- RunKNNPredict(
srt_query = pancreas_sub,
srt_ref = panc8_sub,
ref_group = "celltype",
ref_collapsing = FALSE
)
#> ℹ [2026-02-11 04:03:24] Use the HVF to calculate distance metric
#> ℹ [2026-02-11 04:03:24] Use [1] 632 features to calculate distance.
#> ℹ [2026-02-11 04:03:25] Detected query data type: "log_normalized_counts"
#> ℹ [2026-02-11 04:03:25] Detected reference data type: "log_normalized_counts"
#> ℹ [2026-02-11 04:03:25] Calculate similarity...
#> ℹ [2026-02-11 04:03:25] Use raw method to find neighbors
#> ℹ [2026-02-11 04:03:26] Predict cell type...
CellDimPlot(
pancreas_sub,
group.by = "KNNPredict_classification",
label = TRUE
)
pancreas_sub <- RunKNNPredict(
srt_query = pancreas_sub,
srt_ref = panc8_sub,
ref_group = "celltype",
ref_collapsing = FALSE
)
#> ℹ [2026-02-11 04:03:24] Use the HVF to calculate distance metric
#> ℹ [2026-02-11 04:03:24] Use [1] 632 features to calculate distance.
#> ℹ [2026-02-11 04:03:25] Detected query data type: "log_normalized_counts"
#> ℹ [2026-02-11 04:03:25] Detected reference data type: "log_normalized_counts"
#> ℹ [2026-02-11 04:03:25] Calculate similarity...
#> ℹ [2026-02-11 04:03:25] Use raw method to find neighbors
#> ℹ [2026-02-11 04:03:26] Predict cell type...
CellDimPlot(
pancreas_sub,
group.by = "KNNPredict_classification",
label = TRUE
)
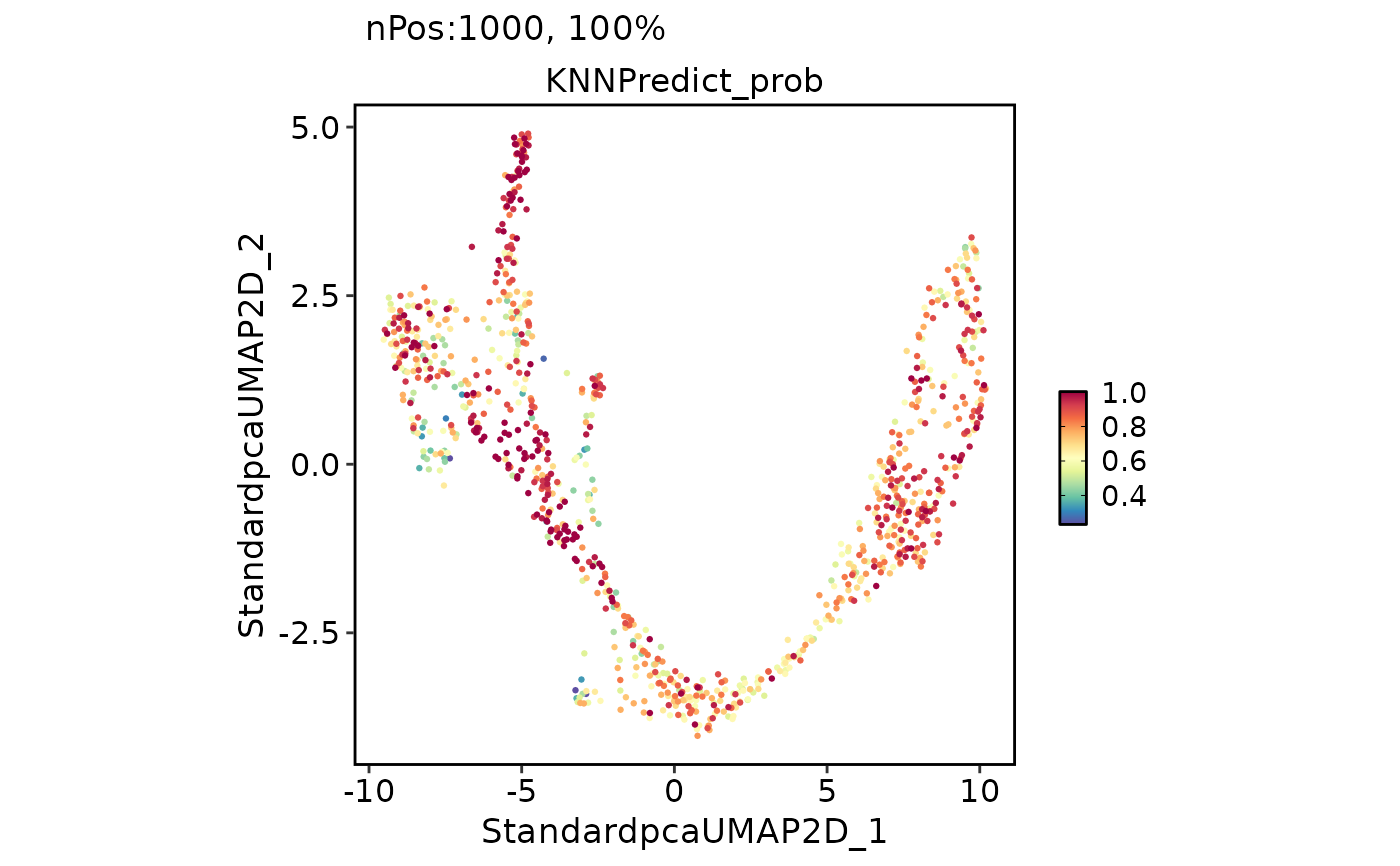
 FeatureDimPlot(
pancreas_sub,
features = "KNNPredict_prob"
)
FeatureDimPlot(
pancreas_sub,
features = "KNNPredict_prob"
)
 pancreas_sub <- RunKNNPredict(
srt_query = pancreas_sub,
srt_ref = panc8_sub,
query_group = "SubCellType",
ref_group = "celltype"
)
#> ℹ [2026-02-11 04:03:26] Use the HVF to calculate distance metric
#> ℹ [2026-02-11 04:03:26] Use [1] 632 features to calculate distance.
#> ℹ [2026-02-11 04:03:26] Detected query data type: "log_normalized_counts"
#> ℹ [2026-02-11 04:03:26] Detected reference data type: "log_normalized_counts"
#> ℹ [2026-02-11 04:03:26] Calculate similarity...
#> ℹ [2026-02-11 04:03:26] Use raw method to find neighbors
#> ℹ [2026-02-11 04:03:26] Predict cell type...
CellDimPlot(
pancreas_sub,
group.by = "KNNPredict_classification",
label = TRUE
)
pancreas_sub <- RunKNNPredict(
srt_query = pancreas_sub,
srt_ref = panc8_sub,
query_group = "SubCellType",
ref_group = "celltype"
)
#> ℹ [2026-02-11 04:03:26] Use the HVF to calculate distance metric
#> ℹ [2026-02-11 04:03:26] Use [1] 632 features to calculate distance.
#> ℹ [2026-02-11 04:03:26] Detected query data type: "log_normalized_counts"
#> ℹ [2026-02-11 04:03:26] Detected reference data type: "log_normalized_counts"
#> ℹ [2026-02-11 04:03:26] Calculate similarity...
#> ℹ [2026-02-11 04:03:26] Use raw method to find neighbors
#> ℹ [2026-02-11 04:03:26] Predict cell type...
CellDimPlot(
pancreas_sub,
group.by = "KNNPredict_classification",
label = TRUE
)
 FeatureDimPlot(
pancreas_sub,
features = "KNNPredict_simil"
)
FeatureDimPlot(
pancreas_sub,
features = "KNNPredict_simil"
)
 # Annotate with DE gene instead of HVF
pancreas_sub <- RunKNNPredict(
srt_query = pancreas_sub,
srt_ref = panc8_sub,
ref_group = "celltype",
features_type = "DE",
feature_source = "ref",
DEtest_param = list(cores = 2)
)
#> ℹ [2026-02-11 04:03:28] Data type is log-normalized
#> ℹ [2026-02-11 04:03:28] Start differential expression test
#> ℹ [2026-02-11 04:03:28] Find all markers(wilcox) among [1] 13 groups...
#> ℹ [2026-02-11 04:03:28] Using 2 cores
#> ⠙ [2026-02-11 04:03:28] Running for delta... [7/13] ■■■■■■■■■■■■■■■■■ …
#> ✔ [2026-02-11 04:03:28] Completed 13 tasks in 4s
#>
#> ℹ [2026-02-11 04:03:28] Building results
#> ✔ [2026-02-11 04:03:32] Differential expression test completed
#> ℹ [2026-02-11 04:03:32] Use the DE features from AllMarkers_wilcox to calculate distance metric.
#> ℹ [2026-02-11 04:03:32] DE features number of the ref data: [1] 1998
#> ℹ [2026-02-11 04:03:32] Use [1] 1998 features to calculate distance.
#> ℹ [2026-02-11 04:03:32] Detected query data type: "log_normalized_counts"
#> ℹ [2026-02-11 04:03:32] Detected reference data type: "log_normalized_counts"
#> ℹ [2026-02-11 04:03:32] Calculate similarity...
#> ℹ [2026-02-11 04:03:32] Use raw method to find neighbors
#> ℹ [2026-02-11 04:03:33] Predict cell type...
CellDimPlot(
pancreas_sub,
group.by = "KNNPredict_classification",
label = TRUE
)
# Annotate with DE gene instead of HVF
pancreas_sub <- RunKNNPredict(
srt_query = pancreas_sub,
srt_ref = panc8_sub,
ref_group = "celltype",
features_type = "DE",
feature_source = "ref",
DEtest_param = list(cores = 2)
)
#> ℹ [2026-02-11 04:03:28] Data type is log-normalized
#> ℹ [2026-02-11 04:03:28] Start differential expression test
#> ℹ [2026-02-11 04:03:28] Find all markers(wilcox) among [1] 13 groups...
#> ℹ [2026-02-11 04:03:28] Using 2 cores
#> ⠙ [2026-02-11 04:03:28] Running for delta... [7/13] ■■■■■■■■■■■■■■■■■ …
#> ✔ [2026-02-11 04:03:28] Completed 13 tasks in 4s
#>
#> ℹ [2026-02-11 04:03:28] Building results
#> ✔ [2026-02-11 04:03:32] Differential expression test completed
#> ℹ [2026-02-11 04:03:32] Use the DE features from AllMarkers_wilcox to calculate distance metric.
#> ℹ [2026-02-11 04:03:32] DE features number of the ref data: [1] 1998
#> ℹ [2026-02-11 04:03:32] Use [1] 1998 features to calculate distance.
#> ℹ [2026-02-11 04:03:32] Detected query data type: "log_normalized_counts"
#> ℹ [2026-02-11 04:03:32] Detected reference data type: "log_normalized_counts"
#> ℹ [2026-02-11 04:03:32] Calculate similarity...
#> ℹ [2026-02-11 04:03:32] Use raw method to find neighbors
#> ℹ [2026-02-11 04:03:33] Predict cell type...
CellDimPlot(
pancreas_sub,
group.by = "KNNPredict_classification",
label = TRUE
)
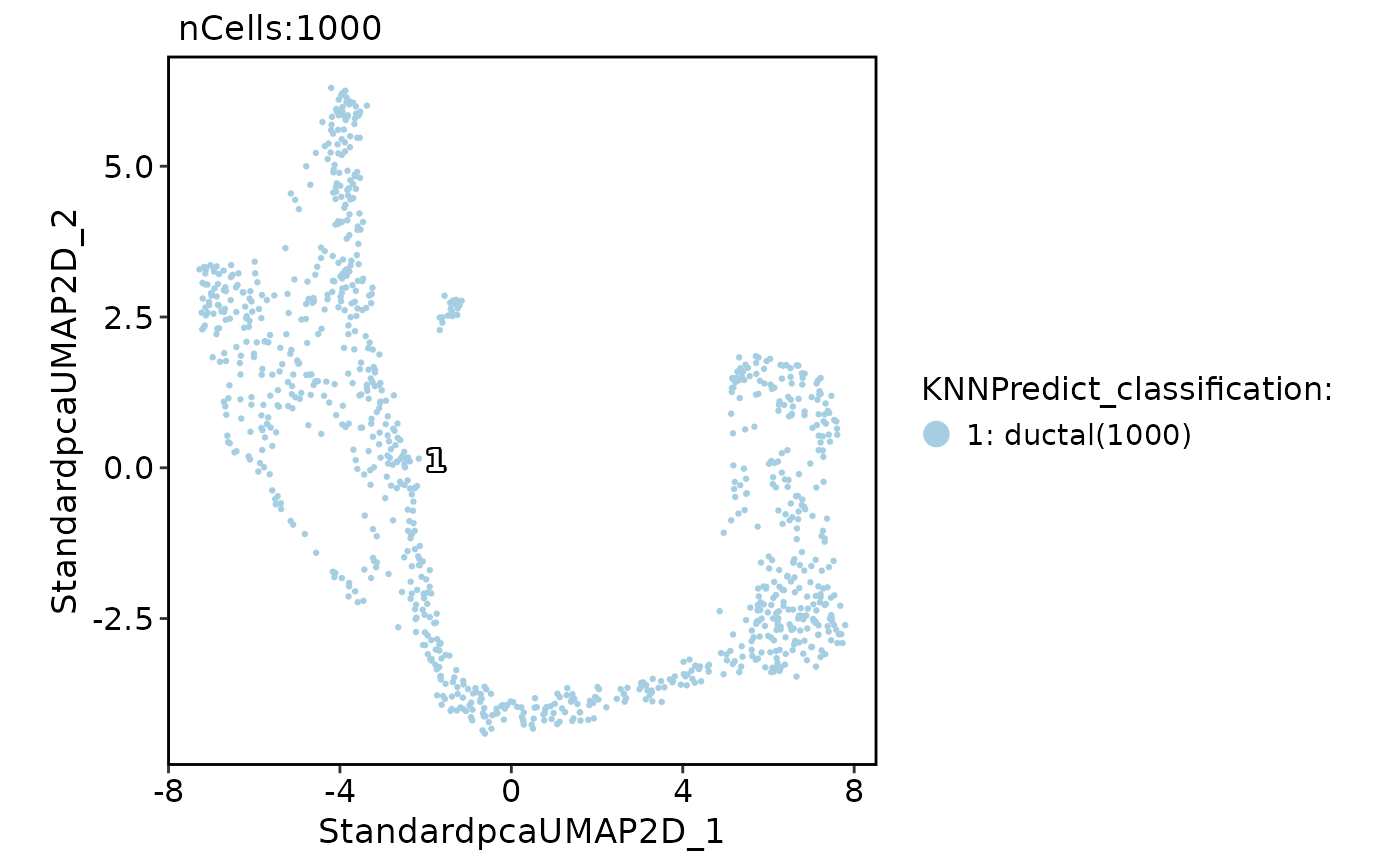 FeatureDimPlot(
pancreas_sub,
features = "KNNPredict_simil"
)
FeatureDimPlot(
pancreas_sub,
features = "KNNPredict_simil"
)
 pancreas_sub <- RunKNNPredict(
srt_query = pancreas_sub,
srt_ref = panc8_sub,
query_group = "SubCellType",
ref_group = "celltype",
features_type = "DE",
feature_source = "both",
DEtest_param = list(cores = 2)
)
#> ℹ [2026-02-11 04:03:34] Data type is log-normalized
#> ℹ [2026-02-11 04:03:34] Start differential expression test
#> ℹ [2026-02-11 04:03:34] Find all markers(wilcox) among [1] 8 groups...
#> ℹ [2026-02-11 04:03:34] Using 2 cores
#> ℹ [2026-02-11 04:03:34] Building results
#> ✔ [2026-02-11 04:03:35] Differential expression test completed
#> ℹ [2026-02-11 04:03:35] Use the DE features from AllMarkers_wilcox to calculate distance metric.
#> ℹ [2026-02-11 04:03:35] DE features number of the query data: [1] 1998
#> ℹ [2026-02-11 04:03:36] Data type is log-normalized
#> ℹ [2026-02-11 04:03:36] Start differential expression test
#> ℹ [2026-02-11 04:03:36] Find all markers(wilcox) among [1] 13 groups...
#> ℹ [2026-02-11 04:03:36] Using 2 cores
#> ⠙ [2026-02-11 04:03:36] Running for delta... [7/13] ■■■■■■■■■■■■■■■■■ …
#> ✔ [2026-02-11 04:03:36] Completed 13 tasks in 3.6s
#>
#> ℹ [2026-02-11 04:03:36] Building results
#> ✔ [2026-02-11 04:03:40] Differential expression test completed
#> ℹ [2026-02-11 04:03:40] Use the DE features from AllMarkers_wilcox to calculate distance metric.
#> ℹ [2026-02-11 04:03:40] DE features number of the ref data: [1] 352
#> ℹ [2026-02-11 04:03:40] Use [1] 102 features to calculate distance.
#> ℹ [2026-02-11 04:03:40] Detected query data type: "log_normalized_counts"
#> ℹ [2026-02-11 04:03:40] Detected reference data type: "log_normalized_counts"
#> ℹ [2026-02-11 04:03:40] Calculate similarity...
#> ℹ [2026-02-11 04:03:40] Use raw method to find neighbors
#> ℹ [2026-02-11 04:03:40] Predict cell type...
CellDimPlot(
pancreas_sub,
group.by = "KNNPredict_classification",
label = TRUE
)
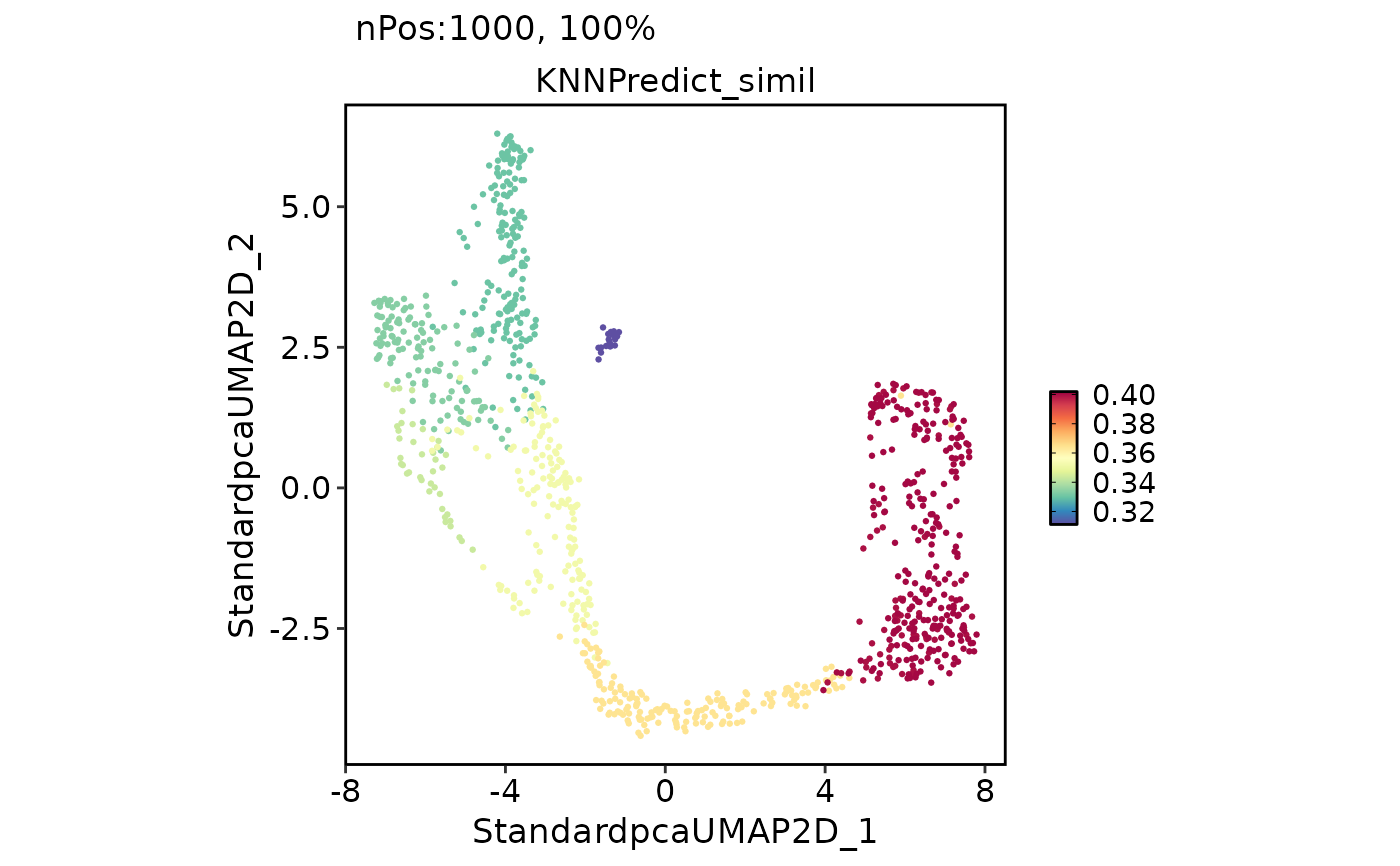
pancreas_sub <- RunKNNPredict(
srt_query = pancreas_sub,
srt_ref = panc8_sub,
query_group = "SubCellType",
ref_group = "celltype",
features_type = "DE",
feature_source = "both",
DEtest_param = list(cores = 2)
)
#> ℹ [2026-02-11 04:03:34] Data type is log-normalized
#> ℹ [2026-02-11 04:03:34] Start differential expression test
#> ℹ [2026-02-11 04:03:34] Find all markers(wilcox) among [1] 8 groups...
#> ℹ [2026-02-11 04:03:34] Using 2 cores
#> ℹ [2026-02-11 04:03:34] Building results
#> ✔ [2026-02-11 04:03:35] Differential expression test completed
#> ℹ [2026-02-11 04:03:35] Use the DE features from AllMarkers_wilcox to calculate distance metric.
#> ℹ [2026-02-11 04:03:35] DE features number of the query data: [1] 1998
#> ℹ [2026-02-11 04:03:36] Data type is log-normalized
#> ℹ [2026-02-11 04:03:36] Start differential expression test
#> ℹ [2026-02-11 04:03:36] Find all markers(wilcox) among [1] 13 groups...
#> ℹ [2026-02-11 04:03:36] Using 2 cores
#> ⠙ [2026-02-11 04:03:36] Running for delta... [7/13] ■■■■■■■■■■■■■■■■■ …
#> ✔ [2026-02-11 04:03:36] Completed 13 tasks in 3.6s
#>
#> ℹ [2026-02-11 04:03:36] Building results
#> ✔ [2026-02-11 04:03:40] Differential expression test completed
#> ℹ [2026-02-11 04:03:40] Use the DE features from AllMarkers_wilcox to calculate distance metric.
#> ℹ [2026-02-11 04:03:40] DE features number of the ref data: [1] 352
#> ℹ [2026-02-11 04:03:40] Use [1] 102 features to calculate distance.
#> ℹ [2026-02-11 04:03:40] Detected query data type: "log_normalized_counts"
#> ℹ [2026-02-11 04:03:40] Detected reference data type: "log_normalized_counts"
#> ℹ [2026-02-11 04:03:40] Calculate similarity...
#> ℹ [2026-02-11 04:03:40] Use raw method to find neighbors
#> ℹ [2026-02-11 04:03:40] Predict cell type...
CellDimPlot(
pancreas_sub,
group.by = "KNNPredict_classification",
label = TRUE
)
 FeatureDimPlot(
pancreas_sub,
features = "KNNPredict_simil"
)
FeatureDimPlot(
pancreas_sub,
features = "KNNPredict_simil"
)