Single-cell reference mapping with Symphony method
Usage
RunSymphonyMap(
srt_query,
srt_ref,
query_assay = NULL,
ref_assay = srt_ref[[ref_pca]]@assay.used,
ref_pca = NULL,
ref_harmony = NULL,
ref_umap = NULL,
ref_group = NULL,
projection_method = c("model", "knn"),
nn_method = NULL,
k = 30,
distance_metric = "cosine",
vote_fun = "mean"
)Arguments
- srt_query
A Seurat object storing the query cells.
- srt_ref
A Seurat object storing the reference cells.
- query_assay
A character string specifying the assay name for the query cells. If not provided, the default assay for the query object will be used.
- ref_assay
A character string specifying the assay name for the reference cells. If not provided, the default assay for the reference object will be used.
- ref_pca
The PCA reduction in the reference object to use for calculating the distance metric.
- ref_harmony
The Harmony reduction in the reference object to use for calculating the distance metric.
- ref_umap
A character string specifying the name of the UMAP reduction in the reference object. If not provided, the first UMAP reduction found in the reference object will be used.
- ref_group
A character string specifying a metadata column name in the reference object to use for grouping.
- projection_method
A character string specifying the projection method to use. Options are "model" and "knn". If "model" is selected, the function will try to use a pre-trained UMAP model in the reference object for projection. If "knn" is selected, the function will directly find the nearest neighbors using the distance metric.
- nn_method
A character string specifying the nearest neighbor search method to use. Options are "raw", "annoy", and "rann". If "raw" is selected, the function will use the brute-force method to find the nearest neighbors. If "annoy" is selected, the function will use the Annoy library for approximate nearest neighbor search. If "rann" is selected, the function will use the RANN library for approximate nearest neighbor search. If not provided, the function will choose the search method based on the size of the query and reference datasets.
- k
A number of nearest neighbors to find for each cell in the query object.
- distance_metric
The distance metric to use for calculating the pairwise distances between cells. Options include: "pearson", "spearman", "cosine", "correlation", "jaccard", "ejaccard", "dice", "edice", "hamman", "simple matching", and "faith". Additional distance metrics can also be used, such as "euclidean", "manhattan", "hamming", etc.
- vote_fun
A character string specifying the function to be used for aggregating the nearest neighbors in the reference object. Options are "mean", "median", "sum", "min", "max", "sd", "var", etc. If not provided, the default is "mean".
Examples
data(panc8_sub)
panc8_sub <- standard_scop(panc8_sub)
#> ℹ [2026-02-11 04:18:21] Start standard scop workflow...
#> ℹ [2026-02-11 04:18:22] Checking a list of <Seurat>...
#> ! [2026-02-11 04:18:22] Data 1/1 of the `srt_list` is "unknown"
#> ℹ [2026-02-11 04:18:22] Perform `NormalizeData()` with `normalization.method = 'LogNormalize'` on the data 1/1 of the `srt_list`...
#> ℹ [2026-02-11 04:18:24] Perform `Seurat::FindVariableFeatures()` on the data 1/1 of the `srt_list`...
#> ℹ [2026-02-11 04:18:25] Use the separate HVF from srt_list
#> ℹ [2026-02-11 04:18:25] Number of available HVF: 2000
#> ℹ [2026-02-11 04:18:25] Finished check
#> ℹ [2026-02-11 04:18:26] Perform `Seurat::ScaleData()`
#> ℹ [2026-02-11 04:18:26] Perform pca linear dimension reduction
#> ℹ [2026-02-11 04:18:29] Perform `Seurat::FindClusters()` with `cluster_algorithm = 'louvain'` and `cluster_resolution = 0.6`
#> ℹ [2026-02-11 04:18:29] Reorder clusters...
#> ℹ [2026-02-11 04:18:29] Perform umap nonlinear dimension reduction
#> ℹ [2026-02-11 04:18:29] Non-linear dimensionality reduction (umap) using (Standardpca) dims (1-50) as input
#> ℹ [2026-02-11 04:18:34] Non-linear dimensionality reduction (umap) using (Standardpca) dims (1-50) as input
#> ✔ [2026-02-11 04:18:39] Run scop standard workflow completed
srt_ref <- panc8_sub[, panc8_sub$tech != "fluidigmc1"]
srt_query <- panc8_sub[, panc8_sub$tech == "fluidigmc1"]
srt_ref <- integration_scop(
srt_ref,
batch = "tech",
integration_method = "Harmony"
)
#> ◌ [2026-02-11 04:18:39] Run Harmony integration...
#> ℹ [2026-02-11 04:18:39] Spliting `srt_merge` into `srt_list` by column "tech"...
#> ℹ [2026-02-11 04:18:40] Checking a list of <Seurat>...
#> ℹ [2026-02-11 04:18:40] Data 1/4 of the `srt_list` has been log-normalized
#> ℹ [2026-02-11 04:18:40] Perform `Seurat::FindVariableFeatures()` on the data 1/4 of the `srt_list`...
#> ℹ [2026-02-11 04:18:41] Data 2/4 of the `srt_list` has been log-normalized
#> ℹ [2026-02-11 04:18:41] Perform `Seurat::FindVariableFeatures()` on the data 2/4 of the `srt_list`...
#> ℹ [2026-02-11 04:18:41] Data 3/4 of the `srt_list` has been log-normalized
#> ℹ [2026-02-11 04:18:41] Perform `Seurat::FindVariableFeatures()` on the data 3/4 of the `srt_list`...
#> ℹ [2026-02-11 04:18:42] Data 4/4 of the `srt_list` has been log-normalized
#> ℹ [2026-02-11 04:18:42] Perform `Seurat::FindVariableFeatures()` on the data 4/4 of the `srt_list`...
#> ℹ [2026-02-11 04:18:42] Use the separate HVF from srt_list
#> ℹ [2026-02-11 04:18:43] Number of available HVF: 2000
#> ℹ [2026-02-11 04:18:43] Finished check
#> ℹ [2026-02-11 04:18:45] Perform `Seurat::ScaleData()`
#> ℹ [2026-02-11 04:18:45] Perform linear dimension reduction("pca")
#> ℹ [2026-02-11 04:18:46] Perform Harmony integration
#> ℹ [2026-02-11 04:18:46] Harmony integration using "Harmonypca", dims:1-10 as input
#> ℹ [2026-02-11 04:18:48] Perform Seurat::FindClusters (louvain)
#> ℹ [2026-02-11 04:18:48] Reorder clusters...
#> ℹ [2026-02-11 04:18:48] Perform nonlinear dimension reduction (umap)
#> ℹ [2026-02-11 04:18:48] Non-linear dimensionality reduction (umap) using (Harmony) dims (1-10) as input
#> ℹ [2026-02-11 04:18:53] Non-linear dimensionality reduction (umap) using (Harmony) dims (1-10) as input
#> ✔ [2026-02-11 04:18:59] Run Harmony integration done
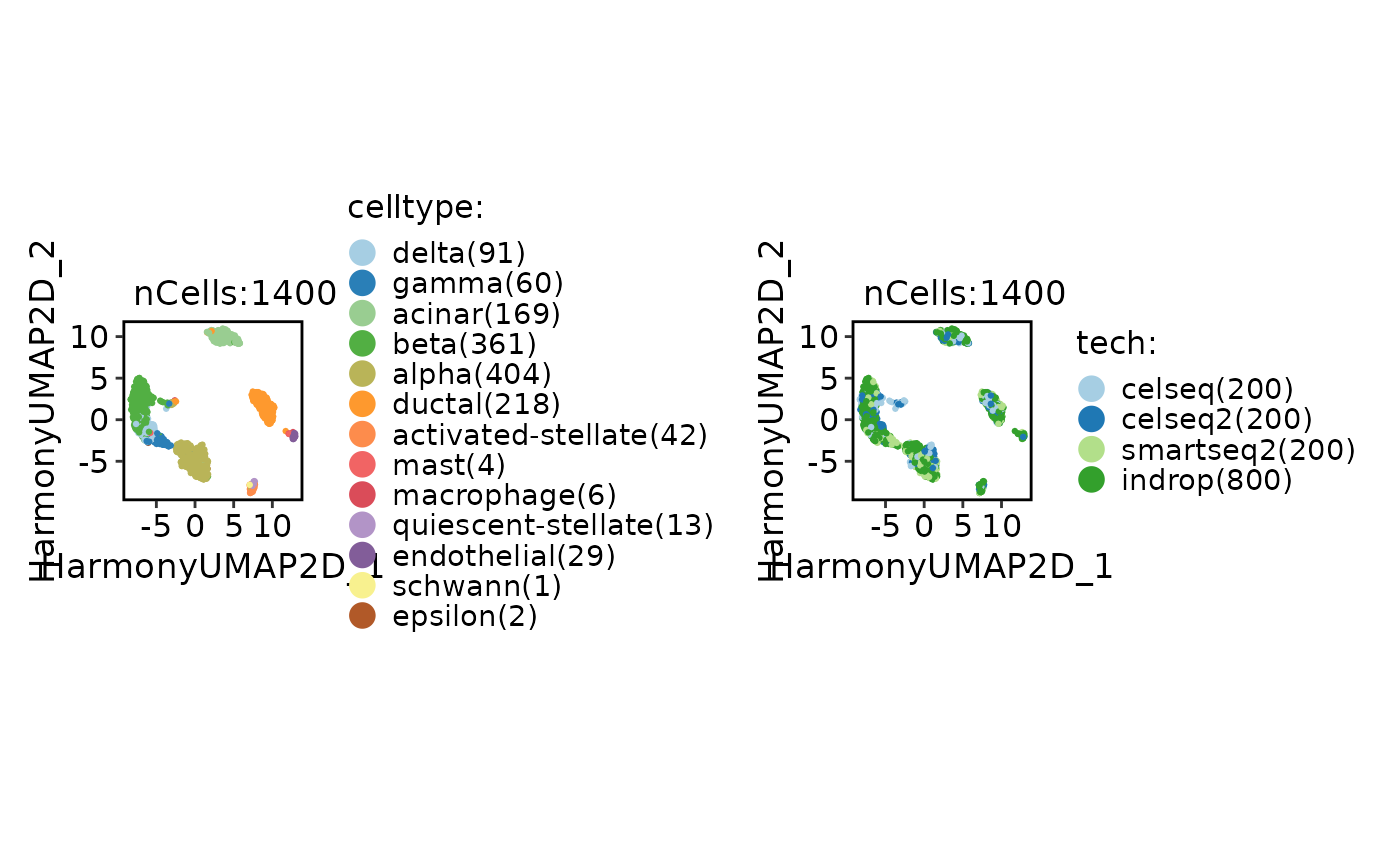
CellDimPlot(srt_ref, group.by = c("celltype", "tech"))
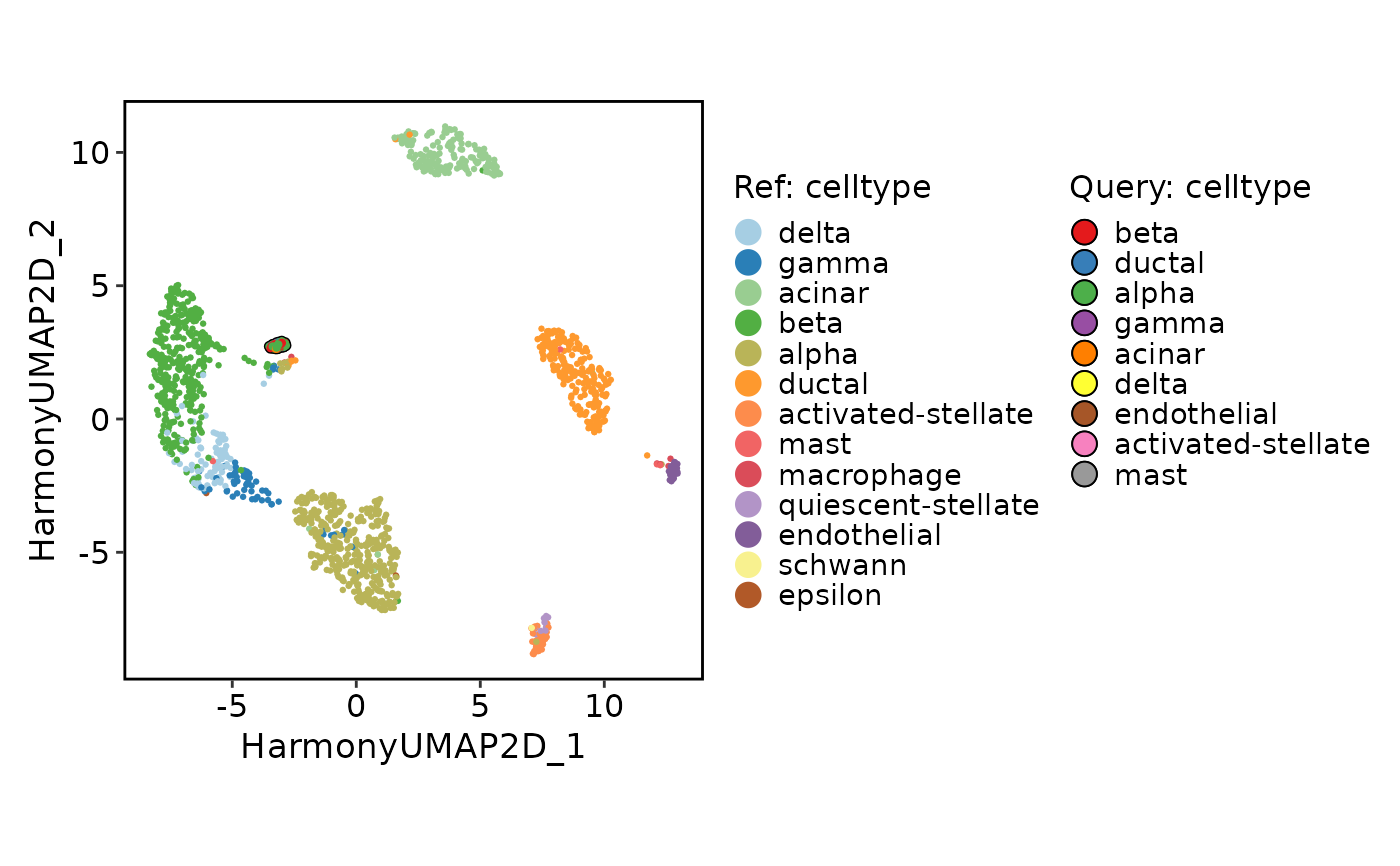
 # Projection
srt_query <- RunSymphonyMap(
srt_query = srt_query,
srt_ref = srt_ref,
ref_pca = "Harmonypca",
ref_harmony = "Harmony",
ref_umap = "HarmonyUMAP2D"
)
#> ℹ [2026-02-11 04:19:29] Data type is log-normalized
#> ℹ [2026-02-11 04:19:29] Detected `srt_query` data type: "log_normalized_counts"
#> ℹ [2026-02-11 04:19:30] Data type is log-normalized
#> ℹ [2026-02-11 04:19:30] Detected `srt_ref` data type: "log_normalized_counts"
#> ℹ [2026-02-11 04:19:30] Build reference
#> ℹ [2026-02-11 04:19:30] Saved embeddings
#> ℹ [2026-02-11 04:19:30] Saved soft cluster assignments
#> ℹ [2026-02-11 04:19:30] Saved variable gene information for 2000 genes
#> ℹ [2026-02-11 04:19:30] Saved PCA loadings
#> ℹ [2026-02-11 04:19:30] Saved metadata
#> ℹ [2026-02-11 04:19:30] Calculate final L2 normalized reference centroids (Y_cos)
#> ℹ [2026-02-11 04:19:30] Calculate reference compression terms (Nr and C)
#> ℹ [2026-02-11 04:19:30] Run mapQuery
#> ℹ [2026-02-11 04:19:30] Scaling and synchronizing query gene expression
#> ℹ [2026-02-11 04:19:30] Found 2000 reference variable genes in query dataset
#> ℹ [2026-02-11 04:19:30] Project query cells using reference gene loadings
#> ℹ [2026-02-11 04:19:30] Clustering query cells to reference centroids
#> ℹ [2026-02-11 04:19:30] Correcting query batch effects
#> ℹ [2026-02-11 04:19:30] Run UMAP projection
#> ℹ [2026-02-11 04:19:30] Use the reduction to calculate distance metric
#> ℹ [2026-02-11 04:19:30] Use raw method to find neighbors
#> ℹ [2026-02-11 04:19:30] Running UMAP projection
#> ℹ [2026-02-11 04:19:31] Run SymphonyMap finished
ProjectionPlot(
srt_query = srt_query,
srt_ref = srt_ref,
query_group = "celltype",
ref_group = "celltype"
)
#> Scale for x is already present.
#> Adding another scale for x, which will replace the existing scale.
#> Scale for y is already present.
#> Adding another scale for y, which will replace the existing scale.
# Projection
srt_query <- RunSymphonyMap(
srt_query = srt_query,
srt_ref = srt_ref,
ref_pca = "Harmonypca",
ref_harmony = "Harmony",
ref_umap = "HarmonyUMAP2D"
)
#> ℹ [2026-02-11 04:19:29] Data type is log-normalized
#> ℹ [2026-02-11 04:19:29] Detected `srt_query` data type: "log_normalized_counts"
#> ℹ [2026-02-11 04:19:30] Data type is log-normalized
#> ℹ [2026-02-11 04:19:30] Detected `srt_ref` data type: "log_normalized_counts"
#> ℹ [2026-02-11 04:19:30] Build reference
#> ℹ [2026-02-11 04:19:30] Saved embeddings
#> ℹ [2026-02-11 04:19:30] Saved soft cluster assignments
#> ℹ [2026-02-11 04:19:30] Saved variable gene information for 2000 genes
#> ℹ [2026-02-11 04:19:30] Saved PCA loadings
#> ℹ [2026-02-11 04:19:30] Saved metadata
#> ℹ [2026-02-11 04:19:30] Calculate final L2 normalized reference centroids (Y_cos)
#> ℹ [2026-02-11 04:19:30] Calculate reference compression terms (Nr and C)
#> ℹ [2026-02-11 04:19:30] Run mapQuery
#> ℹ [2026-02-11 04:19:30] Scaling and synchronizing query gene expression
#> ℹ [2026-02-11 04:19:30] Found 2000 reference variable genes in query dataset
#> ℹ [2026-02-11 04:19:30] Project query cells using reference gene loadings
#> ℹ [2026-02-11 04:19:30] Clustering query cells to reference centroids
#> ℹ [2026-02-11 04:19:30] Correcting query batch effects
#> ℹ [2026-02-11 04:19:30] Run UMAP projection
#> ℹ [2026-02-11 04:19:30] Use the reduction to calculate distance metric
#> ℹ [2026-02-11 04:19:30] Use raw method to find neighbors
#> ℹ [2026-02-11 04:19:30] Running UMAP projection
#> ℹ [2026-02-11 04:19:31] Run SymphonyMap finished
ProjectionPlot(
srt_query = srt_query,
srt_ref = srt_ref,
query_group = "celltype",
ref_group = "celltype"
)
#> Scale for x is already present.
#> Adding another scale for x, which will replace the existing scale.
#> Scale for y is already present.
#> Adding another scale for y, which will replace the existing scale.