This function generates a projection plot, which can be used to compare two groups of cells in a dimensionality reduction space.
Usage
ProjectionPlot(
srt_query,
srt_ref,
query_group = NULL,
ref_group = NULL,
query_reduction = "ref.embeddings",
ref_reduction = srt_query[[query_reduction]]@misc[["reduction.model"]] %||% NULL,
query_param = list(palette = "Set1", cells.highlight = TRUE),
ref_param = list(palette = "Paired"),
xlim = NULL,
ylim = NULL,
pt.size = 0.8,
stroke.highlight = 0.5
)Arguments
- srt_query
A Seurat object storing the query cells.
- srt_ref
A Seurat object storing the reference cells.
- query_group
The grouping variable for the query group cells.
- ref_group
The grouping variable for the reference group cells.
- query_reduction
The name of the reduction in the query group cells.
- ref_reduction
The name of the reduction in the reference group cells.
- query_param
A list of parameters for customizing the query group plot. Available parameters: palette (color palette for groups) and cells.highlight (whether to highlight cells).
- ref_param
A list of parameters for customizing the reference group plot. Available parameters: palette (color palette for groups) and cells.highlight (whether to highlight cells).
- xlim
The x-axis limits for the plot. If not provided, the limits will be calculated based on the data.
- ylim
The y-axis limits for the plot. If not provided, the limits will be calculated based on the data.
- pt.size
The size of the points in the plot.
- stroke.highlight
The size of the stroke highlight for cells.
Examples
data(panc8_sub)
panc8_sub <- standard_scop(panc8_sub)
#> ℹ [2026-02-11 03:37:51] Start standard scop workflow...
#> ℹ [2026-02-11 03:37:52] Checking a list of <Seurat>...
#> ! [2026-02-11 03:37:52] Data 1/1 of the `srt_list` is "unknown"
#> ℹ [2026-02-11 03:37:52] Perform `NormalizeData()` with `normalization.method = 'LogNormalize'` on the data 1/1 of the `srt_list`...
#> ℹ [2026-02-11 03:37:54] Perform `Seurat::FindVariableFeatures()` on the data 1/1 of the `srt_list`...
#> ℹ [2026-02-11 03:37:54] Use the separate HVF from srt_list
#> ℹ [2026-02-11 03:37:55] Number of available HVF: 2000
#> ℹ [2026-02-11 03:37:55] Finished check
#> ℹ [2026-02-11 03:37:55] Perform `Seurat::ScaleData()`
#> ℹ [2026-02-11 03:37:55] Perform pca linear dimension reduction
#> ℹ [2026-02-11 03:37:58] Perform `Seurat::FindClusters()` with `cluster_algorithm = 'louvain'` and `cluster_resolution = 0.6`
#> ℹ [2026-02-11 03:37:58] Reorder clusters...
#> ℹ [2026-02-11 03:37:58] Perform umap nonlinear dimension reduction
#> ℹ [2026-02-11 03:37:58] Non-linear dimensionality reduction (umap) using (Standardpca) dims (1-50) as input
#> ℹ [2026-02-11 03:38:02] Non-linear dimensionality reduction (umap) using (Standardpca) dims (1-50) as input
#> ✔ [2026-02-11 03:38:06] Run scop standard workflow completed
srt_ref <- panc8_sub[, panc8_sub$tech != "fluidigmc1"]
srt_query <- panc8_sub[, panc8_sub$tech == "fluidigmc1"]
srt_ref <- integration_scop(
srt_ref,
batch = "tech",
integration_method = "Uncorrected"
)
#> ◌ [2026-02-11 03:38:06] Run Uncorrected integration...
#> ℹ [2026-02-11 03:38:06] Spliting `srt_merge` into `srt_list` by column "tech"...
#> ℹ [2026-02-11 03:38:07] Checking a list of <Seurat>...
#> ℹ [2026-02-11 03:38:07] Data 1/4 of the `srt_list` has been log-normalized
#> ℹ [2026-02-11 03:38:07] Perform `Seurat::FindVariableFeatures()` on the data 1/4 of the `srt_list`...
#> ℹ [2026-02-11 03:38:08] Data 2/4 of the `srt_list` has been log-normalized
#> ℹ [2026-02-11 03:38:08] Perform `Seurat::FindVariableFeatures()` on the data 2/4 of the `srt_list`...
#> ℹ [2026-02-11 03:38:08] Data 3/4 of the `srt_list` has been log-normalized
#> ℹ [2026-02-11 03:38:08] Perform `Seurat::FindVariableFeatures()` on the data 3/4 of the `srt_list`...
#> ℹ [2026-02-11 03:38:09] Data 4/4 of the `srt_list` has been log-normalized
#> ℹ [2026-02-11 03:38:09] Perform `Seurat::FindVariableFeatures()` on the data 4/4 of the `srt_list`...
#> ℹ [2026-02-11 03:38:09] Use the separate HVF from srt_list
#> ℹ [2026-02-11 03:38:10] Number of available HVF: 2000
#> ℹ [2026-02-11 03:38:10] Finished check
#> ℹ [2026-02-11 03:38:11] Perform Uncorrected integration
#> ℹ [2026-02-11 03:38:12] Perform `Seurat::ScaleData()`
#> ℹ [2026-02-11 03:38:12] Perform linear dimension reduction("pca")
#> ℹ [2026-02-11 03:38:14] Perform Seurat::FindClusters ("louvain")
#> ℹ [2026-02-11 03:38:14] Reorder clusters...
#> ℹ [2026-02-11 03:38:15] Perform nonlinear dimension reduction ("umap")
#> ℹ [2026-02-11 03:38:15] Non-linear dimensionality reduction (umap) using (Uncorrectedpca) dims (1-10) as input
#> ℹ [2026-02-11 03:38:19] Non-linear dimensionality reduction (umap) using (Uncorrectedpca) dims (1-10) as input
#> ✔ [2026-02-11 03:38:24] Run Uncorrected integration done
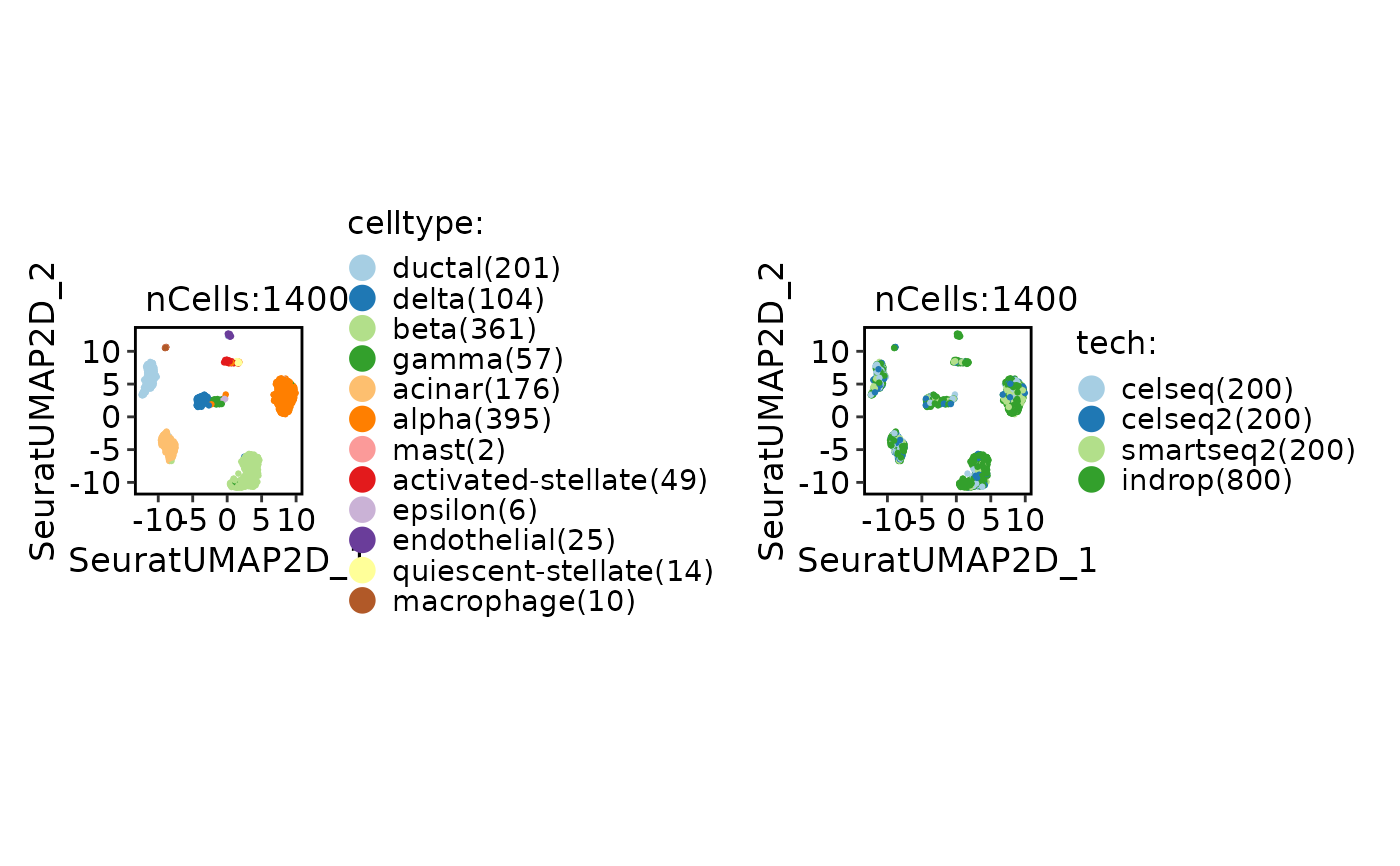
CellDimPlot(
srt_ref,
group.by = c("celltype", "tech")
)
#> Warning: No shared levels found between `names(values)` of the manual scale and the
#> data's fill values.
#> Warning: No shared levels found between `names(values)` of the manual scale and the
#> data's fill values.
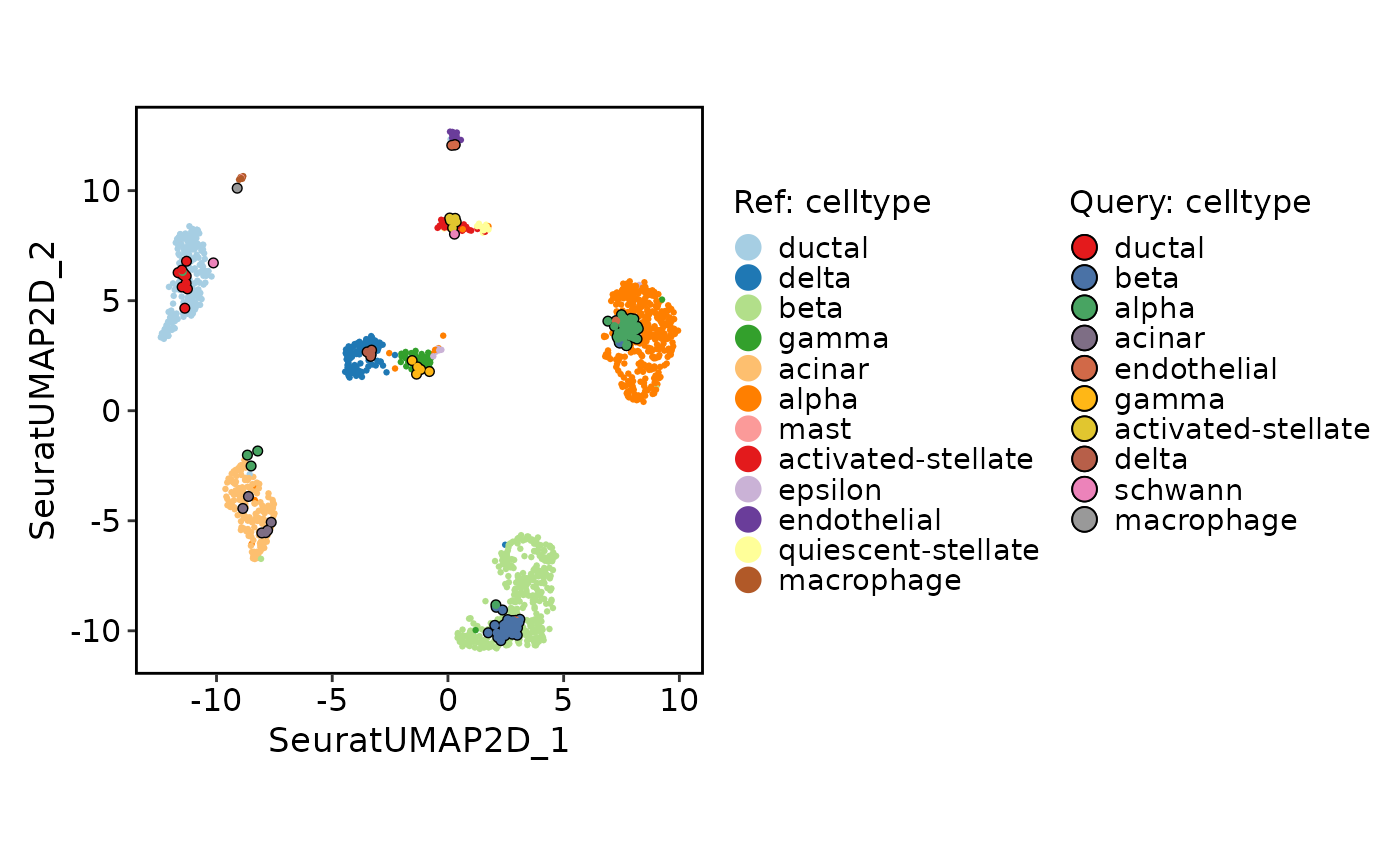
 # Projection
srt_query <- RunKNNMap(
srt_query = srt_query,
srt_ref = srt_ref,
ref_umap = "UncorrectedUMAP2D"
)
#> ℹ [2026-02-11 03:38:24] Use the features to calculate distance metric
#> ℹ [2026-02-11 03:38:24] Data type is log-normalized
#> Warning: The following features were labelled as variable in 'var.features' but had no corresponding rank in `var.features.rank` and will therefore be ignored: 'A4GALT', 'AADAC', 'AARS2', 'ABCA1', 'ABCC3', 'ABCC4', 'ABCC8', 'ABCC9', 'ABCG2', 'ABHD15', 'ABI3', 'ABL2', 'ABTB2', 'ACACB', 'ACBD7', 'ACE', 'ACHE', 'ACSL3', 'ACSL4', 'ACTG1', 'ACTN1', 'ACTN4', 'ADAM12', 'ADAM9', 'ADAMTS1', 'ADAMTS2', 'ADAMTSL2', 'ADAT1', 'ADCK3', 'ADCY1', 'ADCY3', 'ADCY5', 'ADD3', 'ADM', 'ADRA2A', 'ADRA2C', 'ADRB1', 'ADRBK2', 'AFAP1L2', 'AGR2', 'AGRN', 'AHR', 'AIFM3', 'AIM1L', 'AIM1', 'AJAP1', 'AK5', 'AKAP7', 'ALDH1A1', 'ALDH1A2', 'ALDH2', 'ALDH3A2', 'ALDH3B1', 'ALDH4A1', 'ALOX5AP', 'ALPL', 'AMOTL1', 'AMOTL2', 'ANGPTL1', 'ANO6', 'ANO9', 'ANTXR2', 'AOC2', 'AP1S2', 'AP1S3', 'APOBEC2', 'APOD', 'AQP3', 'AQP4', 'ARHGAP26', 'ARHGAP29', 'ARHGAP6', 'ARHGEF17', 'ARHGEF2', 'ARHGEF3', 'ARHGEF40', 'ARHGEF6', 'ARL14', 'ARL4D', 'ARL6', 'ARMC9', 'ARNTL2', 'ARSJ', 'ARX', 'ASAP1', 'ASCL1', 'ASNS', 'ASPH', 'ASRGL1', 'ASS1', 'ASTN2', 'ASXL3', 'ATCAY', 'ATF3', 'ATF5', 'ATHL1', 'ATP10B', 'ATP11A', 'ATP1A1', 'ATP1B2', 'ATP2A3', 'ATP2B4', 'ATP8B1', 'AURKB', 'B3GNT2', 'B4GALT1', 'B4GALT5', 'BACE2', 'BACH2', 'BAIAP2L1', 'BAIAP3', 'BAMBI', 'BARX2', 'BASP1', 'BATF', 'BAZ1A', 'BBC3', 'BCHE', 'BCL2L15', 'BCL3', 'BCL6B', 'BCL6', 'BDH2', 'BHLHE41', 'BHMT2', 'BIK', 'BIRC5', 'BMF', 'BMP2', 'BMPR2', 'BNC2', 'BRIP1', 'BUB1', 'C1QL1', 'C1QTNF1', 'C1RL', 'C2CD4A', 'C2CD4B', 'CABP4', 'CACNA2D1', 'CADM2', 'CADPS2', 'CALCA', 'CALML4', 'CALU', 'CALY', 'CAMK1D', 'CAMKK1', 'CAMKK2', 'CAPN5', 'CAPN6', 'CARHSP1', 'CARTPT', 'CASP6', 'CAV2', 'CBR1', 'CCDC15', 'CCDC71L', 'CCL28', 'CCND2', 'CCNF', 'CCNG2', 'CD200', 'CD276', 'CD47', 'CD68', 'CD82', 'CDCA3', 'CDH17', 'CDH1', 'CDH23', 'CDH2', 'CDH3', 'CDHR2', 'CDK17', 'CDKN1A', 'CDKN2A', 'CDKN2B', 'CDR2', 'CDT1', 'CDX2', 'CEACAM1', 'CEBPB', 'CELF2', 'CENPF', 'CENPW', 'CERCAM', 'CFI', 'CFLAR', 'CGN', 'CHAC1', 'CHGB', 'CHN2', 'CHRM3', 'CHST1', 'CHSY1', 'CITED2', 'CITED4', 'CLCF1', 'CLCN4', 'CLDN11', 'CLDN19', 'CLDN3', 'CLDN5', 'CLDN6', 'CLDN7', 'CLEC11A', 'CLIC5', 'CLIC6', 'CLMN', 'CLMP', 'CLRN3', 'CLSPN', 'CMPK2', 'CMTM3', 'CMTM7', 'CNIH2', 'CNN2', 'CNN3', 'CNNM1', 'CNTN1', 'COCH', 'COL13A1', 'COL14A1', 'COL16A1', 'COL18A1', 'COL5A3', 'COL9A2', 'COMMD2', 'COMP', 'CORO1C', 'CORO2A', 'COX7A1', 'CPA3', 'CPLX2', 'CPM', 'CRABP2', 'CRADD', 'CREB3L1', 'CREB5', 'CREG2', 'CRIP1', 'CRLF1', 'CRTAC1', 'CRTAP', 'CTHRC1', 'CTNNA2', 'CTSB', 'CTSC', 'CTSS', 'CX3CL1', 'CXADR', 'CXCL12', 'CXCL16', 'CYB5A', 'CYBRD1', 'CYP20A1', 'CYP27B1', 'CYR61', 'CYTH3', 'DAB2IP', 'DACH1', 'DACT1', 'DAG1', 'DBN1', 'DCAF11', 'DCBLD2', 'DCUN1D2', 'DDAH2', 'DDC', 'DDIT3', 'DDX25', 'DDX51', 'DGKA', 'DHCR7', 'DHDH', 'DHODH', 'DHRS2', 'DIXDC1', 'DKK1', 'DMBT1', 'DMC1', 'DNAJB9', 'DNAJC22', 'DNER', 'DOCK4', 'DOCK5', 'DOK5', 'DPEP1', 'DPP9', 'DRAM1', 'DSC2', 'DSG2', 'DTNA', 'DUSP10', 'DUSP14', 'DUSP1', 'DUSP23', 'DUSP2', 'DUSP5', 'DZANK1', 'EBP', 'ECE1', 'ECEL1', 'EDN2', 'EFCAB7', 'EFNB1', 'EFNB3', 'EGFR', 'EGF', 'EGR1', 'EGR4', 'EHD1', 'EHD4', 'EHF', 'EIF4EBP1', 'ELAVL4', 'EMILIN1', 'EMP2', 'ENAH', 'ENG', 'ENPP2', 'EPAS1', 'EPDR1', 'EPHA2', 'EPHB2', 'EPHX1', 'EPS8L1', 'ERBB2', 'ERBB3', 'ERO1LB', 'ERO1L', 'ERRFI1', 'ESRRG', 'ETS2', 'ETV1', 'EVA1B', 'EVC2', 'EXT1', 'F10', 'F11R', 'F2RL1', 'F5', 'FA2H', 'FABP5', 'FADS1', 'FADS2', 'FAM101B', 'FAM105A', 'FAM107B', 'FAM129B', 'FAM134B', 'FAM155A', 'FAM159B', 'FAM161A', 'FAM162A', 'FAM163A', 'FAM20C', 'FAM217B', 'FAM227A', 'FAM43A', 'FAM46B', 'FAM46C', 'FAM73A', 'FAM84A', 'FAM84B', 'FBLIM1', 'FBLN1', 'FBLN5', 'FBP1', 'FBXL18', 'FBXO2', 'FBXO6', 'FCER1G', 'FDFT1', 'FERMT2', 'FEV', 'FEZ1', 'FFAR2', 'FFAR4', 'FGD2', 'FGD4', 'FGF7', 'FGFBP1', 'FGFR2', 'FGFR3', 'FGL2', 'FHL2', 'FKBP10', 'FKBP11', 'FKBP5', 'FLNA', 'FNDC4', 'FOLR1', 'FOSB', 'FOSL2', 'FOXC1', 'FOXM1', 'FOXQ1', 'FRAS1', 'FRMD4B', 'FSCN1', 'FXN', 'FXYD3', 'FXYD5', 'FXYD6', 'FZD5', 'G0S2', 'GAD2', 'GADD45A', 'GADD45B', 'GADD45G', 'GALE', 'GALM', 'GALNT2', 'GALNT3', 'GAL', 'GAS6', 'GAS7', 'GATA2', 'GCK', 'GCLC', 'GCNT1', 'GC', 'GDPD1', 'GFOD2', 'GFRA1', 'GGT1', 'GGT5', 'GHR', 'GINS3', 'GINS4', 'GJA1', 'GJA4', 'GJB1', 'GJB3', 'GJC1', 'GK5', 'GK', 'GLDC', 'GLI2', 'GLIPR2', 'GLT1D1', 'GLUL', 'GMDS', 'GMEB1', 'GMFG', 'GMNN', 'GNAL', 'GNPNAT1', 'GNPTAB', 'GOLM1', 'GOT1', 'GPC1', 'GPC6', 'GPR155', 'GPR160', 'GPR161', 'GPR4', 'GPRC5A', 'GPRC5B', 'GPSM1', 'GPT2', 'GPX3', 'GREB1', 'GSN', 'GSTM3', 'GSTM5', 'GSTP1', 'GTF2A1', 'GTPBP10', 'GUCA1B', 'GUCA2A', 'GUCY1A3', 'GULP1', 'GYLTL1B', 'H19', 'HABP2', 'HAP1', 'HBEGF', 'HCK', 'HDHD3', 'HEATR5A', 'HEBP1', 'HEG1', 'HEPH', 'HERPUD1', 'HES1', 'HIC1', 'HIF1A', 'HILPDA', 'HIP1', 'HIRA', 'HIST1H1C', 'HIST1H2BK', 'HIST2H2BE', 'HK1', 'HK2', 'HKDC1', 'HMGB3', 'HMGCS1', 'HMGN5', 'HMOX1', 'HN1', 'HNF1B', 'HNF4A', 'HNMT', 'HOMER2', 'HOPX', 'HOXB2', 'HOXB4', 'HPCAL1', 'HPGD', 'HPN', 'HS3ST1', 'HS3ST3A1', 'HS6ST2', 'HSD11B2', 'HSD17B11', 'HSPA1A', 'HSPA1B', 'HSPA5', 'HSPB6', 'HSPB8', 'HTRA1', 'HUNK', 'HYAL3', 'IBA57', 'ID1', 'ID4', 'IDH1', 'IDH2', 'IER2', 'IER3', 'IFI30', 'IFIT1', 'IFITM2', 'IFNGR1', 'IGFBP6', 'IGFBPL1', 'IGSF3', 'IKBIP', 'IKBKE', 'IKZF3', 'IL11', 'IL15RA', 'IL1RN', 'IL22RA1', 'IL33', 'IMPA2', 'INHBB', 'INPP4B', 'INSIG1', 'INSM1', 'IQGAP2', 'IRAK2', 'IRF1', 'IRS2', 'IRX1', 'IRX2', 'ISG20', 'ISL1', 'ITGA11', 'ITGA1', 'ITGA3', 'ITGA6', 'ITGAV', 'ITGB1', 'ITGB4', 'ITGB8', 'ITIH5', 'ITPR2', 'IVNS1ABP', 'IYD', 'JAM3', 'JARID2', 'JDP2', 'JMJD6', 'JPX', 'JUNB', 'JUN', 'JUP', 'KCNA5', 'KCNE3', 'KCNG1', 'KCNH6', 'KCNJ13', 'KCNJ5', 'KCNJ6', 'KCNJ8', 'KCNK16', 'KCNK5', 'KCNN2', 'KCNN3', 'KCNQ1OT1', 'KCNQ1', 'KCTD12', 'KDELC2', 'KDELR3', 'KIF11', 'KIF12', 'KIF20B', 'KIF21B', 'KIF5C', 'KIT', 'KLB', 'KLF2', 'KLF4', 'KLF5', 'KLF6', 'KLF7', 'KLHL28', 'KLHL31', 'KLHL3', 'KLK10', 'KLK11', 'KREMEN1', 'KRT15', 'KRT17', 'KRTCAP3', 'KYNU', 'L1TD1', 'L2HGDH', 'LAD1', 'LAIR1', 'LAMA1', 'LAMA3', 'LAMB1', 'LAMB2', 'LAMB3', 'LARP6', 'LASP1', 'LBH', 'LCP1', 'LDB2', 'LDLR', 'LEFTY1', 'LFNG', 'LGALS3', 'LGALS9', 'LHFPL2', 'LIFR', 'LIF', 'LIMA1', 'LIMK2', 'LIMS1', 'LIPC', 'LIPH', 'LITAF', 'LMO1', 'LMO2', 'LMO4', 'LMO7', 'LNX2', 'LOXL1', 'LOXL2', 'LPAR1', 'LRP1', 'LRP8', 'LRRC27', 'LRRC2', 'LRRC57', 'LRRC58', 'LRRIQ1', 'LRRK2', 'LRRTM2', 'LSR', 'LSS', 'LTBP4', 'LTB', 'LTF', 'LTV1', 'LURAP1L', 'LXN', 'LY6E', 'LY6H', 'LY96', 'LYN', 'LYPD1', 'LYRM7', 'LZTS1', 'MAB21L3', 'MACC1', 'MAFB', 'MAFF', 'MAL2', 'MALL', 'MAOA', 'MAOB', 'MAP3K9', 'MAPT', 'MARCKSL1', 'MAT1A', 'MCC', 'MCL1', 'MCM2', 'MCM6', 'MCOLN2', 'MDFIC', 'MDFI', 'MDGA1', 'MDK', 'MDM1', 'ME1', 'MEDAG', 'MEF2C', 'MEIS2', 'METRNL', 'METTL21A', 'MEX3A', 'MFAP2', 'MFAP4', 'MFGE8', 'MGAT4B', 'MGLL', 'MGP', 'MICAL2', 'MID1', 'MIR143HG', 'MKNK1', 'MLLT11', 'MLXIPL', 'MMACHC', 'MMP14', 'MNS1', 'MOB3B', 'MOSPD1', 'MOXD1', 'MPPED2', 'MPST', 'MPZL2', 'MRC2', 'MS4A2', 'MSMO1', 'MSRB1', 'MSRB3', 'MSX1', 'MTHFD2', 'MTUS1', 'MVP', 'MYADM', 'MYH9', 'MYL12A', 'MYL12B', 'MYL9', 'MYLK', 'MYO1C', 'MYO5B', 'MYOF', 'MYRF', 'MYZAP', 'N4BP3', 'NAA40', 'NAP1L2', 'NCAM1', 'NCF2', 'NCMAP', 'NCS1', 'NDRG1', 'NDRG2', 'NDUFA4L2', 'NDUFA4', 'NEDD9', 'NEK6', 'NEK8', 'NEURL3', 'NEUROD1', 'NEXN', 'NFATC4', 'NFKBIE', 'NFKBIZ', 'NHSL1', 'NICN1', 'NID1', 'NID2', 'NIPSNAP3B', 'NKD1', 'NLN', 'NLRC3', 'NMB', 'NOL9', 'NOTCH1', 'NOTCH3', 'NOTCH4', 'NPHS1', 'NPR1', 'NPTX2', 'NPY1R', 'NQO1', 'NR0B1', 'NR0B2', 'NR1D1', 'NR2F2', 'NR4A2', 'NR4A3', 'NR5A2', 'NR6A1', 'NRARP', 'NREP', 'NRP2', 'NSG1', 'NT5E', 'NTM', 'NTN4', 'NUAK2', 'NUGGC', 'NUSAP1', 'NYAP2', 'OAF', 'OAS3', 'OCLN', 'ODF2L', 'OGDH', 'OLFM2', 'OLFML1', 'OLFML2B', 'ORAI2', 'ORC4', 'OSGIN1', 'OSMR', 'OVOL1', 'OXR1', 'P2RX1', 'P4HA1', 'P4HA3', 'PACSIN2', 'PAG1', 'PAH', 'PALB2', 'PAMR1', 'PANK1', 'PAQR5', 'PARD6G', 'PARM1', 'PASK', 'PCDH10', 'PCDH17', 'PCDH18', 'PCDH7', 'PCK2', 'PCP4', 'PCSK2', 'PCSK5', 'PDCD4', 'PDDC1', 'PDE1A', 'PDE3B', 'PDE4A', 'PDE4C', 'PDE7B', 'PDE8B', 'PDGFA', 'PDGFB', 'PDGFC', 'PDIA4', 'PDLIM1', 'PDLIM2', 'PDLIM3', 'PDLIM4', 'PDP1', 'PDP2', 'PDX1', 'PEAK1', 'PECAM1', 'PEG10', 'PEMT', 'PGC', 'PGM1', 'PGM2L1', 'PHGDH', 'PHGR1', 'PHLDA1', 'PHLDA2', 'PHLDA3', 'PID1', 'PIEZO1', 'PIGN', 'PIK3IP1', 'PIK3R1', 'PIM1', 'PIM2', 'PIM3', 'PIWIL2', 'PKIB', 'PLA2G15', 'PLA2G4A', 'PLA2G4C', 'PLAT', 'PLBD1', 'PLCD1', 'PLCD3', 'P
#> ℹ [2026-02-11 03:38:26] Data type is log-normalized
#> Warning: The following features were labelled as variable in 'var.features' but had no corresponding rank in `var.features.rank` and will therefore be ignored: 'A4GALT', 'AADAC', 'AARS2', 'ABCA1', 'ABCC3', 'ABCC4', 'ABCC8', 'ABCC9', 'ABCG2', 'ABHD15', 'ABI3', 'ABL2', 'ABTB2', 'ACACB', 'ACBD7', 'ACE', 'ACHE', 'ACSL3', 'ACSL4', 'ACTG1', 'ACTN1', 'ACTN4', 'ADAM12', 'ADAM9', 'ADAMTS1', 'ADAMTS2', 'ADAMTSL2', 'ADAT1', 'ADCK3', 'ADCY1', 'ADCY3', 'ADCY5', 'ADD3', 'ADM', 'ADRA2A', 'ADRA2C', 'ADRB1', 'ADRBK2', 'AFAP1L2', 'AGR2', 'AGRN', 'AHR', 'AIFM3', 'AIM1L', 'AIM1', 'AJAP1', 'AK5', 'AKAP7', 'ALDH1A1', 'ALDH1A2', 'ALDH2', 'ALDH3A2', 'ALDH3B1', 'ALDH4A1', 'ALOX5AP', 'ALPL', 'AMOTL1', 'AMOTL2', 'ANGPTL1', 'ANO6', 'ANO9', 'ANTXR2', 'AOC2', 'AP1S2', 'AP1S3', 'APOBEC2', 'APOD', 'AQP3', 'AQP4', 'ARHGAP26', 'ARHGAP29', 'ARHGAP6', 'ARHGEF17', 'ARHGEF2', 'ARHGEF3', 'ARHGEF40', 'ARHGEF6', 'ARL14', 'ARL4D', 'ARL6', 'ARMC9', 'ARNTL2', 'ARSJ', 'ARX', 'ASAP1', 'ASCL1', 'ASNS', 'ASPH', 'ASRGL1', 'ASS1', 'ASTN2', 'ASXL3', 'ATCAY', 'ATF3', 'ATF5', 'ATHL1', 'ATP10B', 'ATP11A', 'ATP1A1', 'ATP1B2', 'ATP2A3', 'ATP2B4', 'ATP8B1', 'AURKB', 'B3GNT2', 'B4GALT1', 'B4GALT5', 'BACE2', 'BACH2', 'BAIAP2L1', 'BAIAP3', 'BAMBI', 'BARX2', 'BASP1', 'BATF', 'BAZ1A', 'BBC3', 'BCHE', 'BCL2L15', 'BCL3', 'BCL6B', 'BCL6', 'BDH2', 'BHLHE41', 'BHMT2', 'BIK', 'BIRC5', 'BMF', 'BMP2', 'BMPR2', 'BNC2', 'BRIP1', 'BUB1', 'C1QL1', 'C1QTNF1', 'C1RL', 'C2CD4A', 'C2CD4B', 'CABP4', 'CACNA2D1', 'CADM2', 'CADPS2', 'CALCA', 'CALML4', 'CALU', 'CALY', 'CAMK1D', 'CAMKK1', 'CAMKK2', 'CAPN5', 'CAPN6', 'CARHSP1', 'CARTPT', 'CASP6', 'CAV2', 'CBR1', 'CCDC15', 'CCDC71L', 'CCL28', 'CCND2', 'CCNF', 'CCNG2', 'CD200', 'CD276', 'CD47', 'CD68', 'CD82', 'CDCA3', 'CDH17', 'CDH1', 'CDH23', 'CDH2', 'CDH3', 'CDHR2', 'CDK17', 'CDKN1A', 'CDKN2A', 'CDKN2B', 'CDR2', 'CDT1', 'CDX2', 'CEACAM1', 'CEBPB', 'CELF2', 'CENPF', 'CENPW', 'CERCAM', 'CFI', 'CFLAR', 'CGN', 'CHAC1', 'CHGB', 'CHN2', 'CHRM3', 'CHST1', 'CHSY1', 'CITED2', 'CITED4', 'CLCF1', 'CLCN4', 'CLDN11', 'CLDN19', 'CLDN3', 'CLDN5', 'CLDN6', 'CLDN7', 'CLEC11A', 'CLIC5', 'CLIC6', 'CLMN', 'CLMP', 'CLRN3', 'CLSPN', 'CMPK2', 'CMTM3', 'CMTM7', 'CNIH2', 'CNN2', 'CNN3', 'CNNM1', 'CNTN1', 'COCH', 'COL13A1', 'COL14A1', 'COL16A1', 'COL18A1', 'COL5A3', 'COL9A2', 'COMMD2', 'COMP', 'CORO1C', 'CORO2A', 'COX7A1', 'CPA3', 'CPLX2', 'CPM', 'CRABP2', 'CRADD', 'CREB3L1', 'CREB5', 'CREG2', 'CRIP1', 'CRLF1', 'CRTAC1', 'CRTAP', 'CTHRC1', 'CTNNA2', 'CTSB', 'CTSC', 'CTSS', 'CX3CL1', 'CXADR', 'CXCL12', 'CXCL16', 'CYB5A', 'CYBRD1', 'CYP20A1', 'CYP27B1', 'CYR61', 'CYTH3', 'DAB2IP', 'DACH1', 'DACT1', 'DAG1', 'DBN1', 'DCAF11', 'DCBLD2', 'DCUN1D2', 'DDAH2', 'DDC', 'DDIT3', 'DDX25', 'DDX51', 'DGKA', 'DHCR7', 'DHDH', 'DHODH', 'DHRS2', 'DIXDC1', 'DKK1', 'DMBT1', 'DMC1', 'DNAJB9', 'DNAJC22', 'DNER', 'DOCK4', 'DOCK5', 'DOK5', 'DPEP1', 'DPP9', 'DRAM1', 'DSC2', 'DSG2', 'DTNA', 'DUSP10', 'DUSP14', 'DUSP1', 'DUSP23', 'DUSP2', 'DUSP5', 'DZANK1', 'EBP', 'ECE1', 'ECEL1', 'EDN2', 'EFCAB7', 'EFNB1', 'EFNB3', 'EGFR', 'EGF', 'EGR1', 'EGR4', 'EHD1', 'EHD4', 'EHF', 'EIF4EBP1', 'ELAVL4', 'EMILIN1', 'EMP2', 'ENAH', 'ENG', 'ENPP2', 'EPAS1', 'EPDR1', 'EPHA2', 'EPHB2', 'EPHX1', 'EPS8L1', 'ERBB2', 'ERBB3', 'ERO1LB', 'ERO1L', 'ERRFI1', 'ESRRG', 'ETS2', 'ETV1', 'EVA1B', 'EVC2', 'EXT1', 'F10', 'F11R', 'F2RL1', 'F5', 'FA2H', 'FABP5', 'FADS1', 'FADS2', 'FAM101B', 'FAM105A', 'FAM107B', 'FAM129B', 'FAM134B', 'FAM155A', 'FAM159B', 'FAM161A', 'FAM162A', 'FAM163A', 'FAM20C', 'FAM217B', 'FAM227A', 'FAM43A', 'FAM46B', 'FAM46C', 'FAM73A', 'FAM84A', 'FAM84B', 'FBLIM1', 'FBLN1', 'FBLN5', 'FBP1', 'FBXL18', 'FBXO2', 'FBXO6', 'FCER1G', 'FDFT1', 'FERMT2', 'FEV', 'FEZ1', 'FFAR2', 'FFAR4', 'FGD2', 'FGD4', 'FGF7', 'FGFBP1', 'FGFR2', 'FGFR3', 'FGL2', 'FHL2', 'FKBP10', 'FKBP11', 'FKBP5', 'FLNA', 'FNDC4', 'FOLR1', 'FOSB', 'FOSL2', 'FOXC1', 'FOXM1', 'FOXQ1', 'FRAS1', 'FRMD4B', 'FSCN1', 'FXN', 'FXYD3', 'FXYD5', 'FXYD6', 'FZD5', 'G0S2', 'GAD2', 'GADD45A', 'GADD45B', 'GADD45G', 'GALE', 'GALM', 'GALNT2', 'GALNT3', 'GAL', 'GAS6', 'GAS7', 'GATA2', 'GCK', 'GCLC', 'GCNT1', 'GC', 'GDPD1', 'GFOD2', 'GFRA1', 'GGT1', 'GGT5', 'GHR', 'GINS3', 'GINS4', 'GJA1', 'GJA4', 'GJB1', 'GJB3', 'GJC1', 'GK5', 'GK', 'GLDC', 'GLI2', 'GLIPR2', 'GLT1D1', 'GLUL', 'GMDS', 'GMEB1', 'GMFG', 'GMNN', 'GNAL', 'GNPNAT1', 'GNPTAB', 'GOLM1', 'GOT1', 'GPC1', 'GPC6', 'GPR155', 'GPR160', 'GPR161', 'GPR4', 'GPRC5A', 'GPRC5B', 'GPSM1', 'GPT2', 'GPX3', 'GREB1', 'GSN', 'GSTM3', 'GSTM5', 'GSTP1', 'GTF2A1', 'GTPBP10', 'GUCA1B', 'GUCA2A', 'GUCY1A3', 'GULP1', 'GYLTL1B', 'H19', 'HABP2', 'HAP1', 'HBEGF', 'HCK', 'HDHD3', 'HEATR5A', 'HEBP1', 'HEG1', 'HEPH', 'HERPUD1', 'HES1', 'HIC1', 'HIF1A', 'HILPDA', 'HIP1', 'HIRA', 'HIST1H1C', 'HIST1H2BK', 'HIST2H2BE', 'HK1', 'HK2', 'HKDC1', 'HMGB3', 'HMGCS1', 'HMGN5', 'HMOX1', 'HN1', 'HNF1B', 'HNF4A', 'HNMT', 'HOMER2', 'HOPX', 'HOXB2', 'HOXB4', 'HPCAL1', 'HPGD', 'HPN', 'HS3ST1', 'HS3ST3A1', 'HS6ST2', 'HSD11B2', 'HSD17B11', 'HSPA1A', 'HSPA1B', 'HSPA5', 'HSPB6', 'HSPB8', 'HTRA1', 'HUNK', 'HYAL3', 'IBA57', 'ID1', 'ID4', 'IDH1', 'IDH2', 'IER2', 'IER3', 'IFI30', 'IFIT1', 'IFITM2', 'IFNGR1', 'IGFBP6', 'IGFBPL1', 'IGSF3', 'IKBIP', 'IKBKE', 'IKZF3', 'IL11', 'IL15RA', 'IL1RN', 'IL22RA1', 'IL33', 'IMPA2', 'INHBB', 'INPP4B', 'INSIG1', 'INSM1', 'IQGAP2', 'IRAK2', 'IRF1', 'IRS2', 'IRX1', 'IRX2', 'ISG20', 'ISL1', 'ITGA11', 'ITGA1', 'ITGA3', 'ITGA6', 'ITGAV', 'ITGB1', 'ITGB4', 'ITGB8', 'ITIH5', 'ITPR2', 'IVNS1ABP', 'IYD', 'JAM3', 'JARID2', 'JDP2', 'JMJD6', 'JPX', 'JUNB', 'JUN', 'JUP', 'KCNA5', 'KCNE3', 'KCNG1', 'KCNH6', 'KCNJ13', 'KCNJ5', 'KCNJ6', 'KCNJ8', 'KCNK16', 'KCNK5', 'KCNN2', 'KCNN3', 'KCNQ1OT1', 'KCNQ1', 'KCTD12', 'KDELC2', 'KDELR3', 'KIF11', 'KIF12', 'KIF20B', 'KIF21B', 'KIF5C', 'KIT', 'KLB', 'KLF2', 'KLF4', 'KLF5', 'KLF6', 'KLF7', 'KLHL28', 'KLHL31', 'KLHL3', 'KLK10', 'KLK11', 'KREMEN1', 'KRT15', 'KRT17', 'KRTCAP3', 'KYNU', 'L1TD1', 'L2HGDH', 'LAD1', 'LAIR1', 'LAMA1', 'LAMA3', 'LAMB1', 'LAMB2', 'LAMB3', 'LARP6', 'LASP1', 'LBH', 'LCP1', 'LDB2', 'LDLR', 'LEFTY1', 'LFNG', 'LGALS3', 'LGALS9', 'LHFPL2', 'LIFR', 'LIF', 'LIMA1', 'LIMK2', 'LIMS1', 'LIPC', 'LIPH', 'LITAF', 'LMO1', 'LMO2', 'LMO4', 'LMO7', 'LNX2', 'LOXL1', 'LOXL2', 'LPAR1', 'LRP1', 'LRP8', 'LRRC27', 'LRRC2', 'LRRC57', 'LRRC58', 'LRRIQ1', 'LRRK2', 'LRRTM2', 'LSR', 'LSS', 'LTBP4', 'LTB', 'LTF', 'LTV1', 'LURAP1L', 'LXN', 'LY6E', 'LY6H', 'LY96', 'LYN', 'LYPD1', 'LYRM7', 'LZTS1', 'MAB21L3', 'MACC1', 'MAFB', 'MAFF', 'MAL2', 'MALL', 'MAOA', 'MAOB', 'MAP3K9', 'MAPT', 'MARCKSL1', 'MAT1A', 'MCC', 'MCL1', 'MCM2', 'MCM6', 'MCOLN2', 'MDFIC', 'MDFI', 'MDGA1', 'MDK', 'MDM1', 'ME1', 'MEDAG', 'MEF2C', 'MEIS2', 'METRNL', 'METTL21A', 'MEX3A', 'MFAP2', 'MFAP4', 'MFGE8', 'MGAT4B', 'MGLL', 'MGP', 'MICAL2', 'MID1', 'MIR143HG', 'MKNK1', 'MLLT11', 'MLXIPL', 'MMACHC', 'MMP14', 'MNS1', 'MOB3B', 'MOSPD1', 'MOXD1', 'MPPED2', 'MPST', 'MPZL2', 'MRC2', 'MS4A2', 'MSMO1', 'MSRB1', 'MSRB3', 'MSX1', 'MTHFD2', 'MTUS1', 'MVP', 'MYADM', 'MYH9', 'MYL12A', 'MYL12B', 'MYL9', 'MYLK', 'MYO1C', 'MYO5B', 'MYOF', 'MYRF', 'MYZAP', 'N4BP3', 'NAA40', 'NAP1L2', 'NCAM1', 'NCF2', 'NCMAP', 'NCS1', 'NDRG1', 'NDRG2', 'NDUFA4L2', 'NDUFA4', 'NEDD9', 'NEK6', 'NEK8', 'NEURL3', 'NEUROD1', 'NEXN', 'NFATC4', 'NFKBIE', 'NFKBIZ', 'NHSL1', 'NICN1', 'NID1', 'NID2', 'NIPSNAP3B', 'NKD1', 'NLN', 'NLRC3', 'NMB', 'NOL9', 'NOTCH1', 'NOTCH3', 'NOTCH4', 'NPHS1', 'NPR1', 'NPTX2', 'NPY1R', 'NQO1', 'NR0B1', 'NR0B2', 'NR1D1', 'NR2F2', 'NR4A2', 'NR4A3', 'NR5A2', 'NR6A1', 'NRARP', 'NREP', 'NRP2', 'NSG1', 'NT5E', 'NTM', 'NTN4', 'NUAK2', 'NUGGC', 'NUSAP1', 'NYAP2', 'OAF', 'OAS3', 'OCLN', 'ODF2L', 'OGDH', 'OLFM2', 'OLFML1', 'OLFML2B', 'ORAI2', 'ORC4', 'OSGIN1', 'OSMR', 'OVOL1', 'OXR1', 'P2RX1', 'P4HA1', 'P4HA3', 'PACSIN2', 'PAG1', 'PAH', 'PALB2', 'PAMR1', 'PANK1', 'PAQR5', 'PARD6G', 'PARM1', 'PASK', 'PCDH10', 'PCDH17', 'PCDH18', 'PCDH7', 'PCK2', 'PCP4', 'PCSK2', 'PCSK5', 'PDCD4', 'PDDC1', 'PDE1A', 'PDE3B', 'PDE4A', 'PDE4C', 'PDE7B', 'PDE8B', 'PDGFA', 'PDGFB', 'PDGFC', 'PDIA4', 'PDLIM1', 'PDLIM2', 'PDLIM3', 'PDLIM4', 'PDP1', 'PDP2', 'PDX1', 'PEAK1', 'PECAM1', 'PEG10', 'PEMT', 'PGC', 'PGM1', 'PGM2L1', 'PHGDH', 'PHGR1', 'PHLDA1', 'PHLDA2', 'PHLDA3', 'PID1', 'PIEZO1', 'PIGN', 'PIK3IP1', 'PIK3R1', 'PIM1', 'PIM2', 'PIM3', 'PIWIL2', 'PKIB', 'PLA2G15', 'PLA2G4A', 'PLA2G4C', 'PLAT', 'PLBD1', 'PLCD1', 'PLCD3', 'P
#> ℹ [2026-02-11 03:38:26] Use 2000 features to calculate distance
#> Warning: The following features were labelled as variable in 'var.features' but had no corresponding rank in `var.features.rank` and will therefore be ignored: 'A4GALT', 'AADAC', 'AARS2', 'ABCA1', 'ABCC3', 'ABCC4', 'ABCC8', 'ABCC9', 'ABCG2', 'ABHD15', 'ABI3', 'ABL2', 'ABTB2', 'ACACB', 'ACBD7', 'ACE', 'ACHE', 'ACSL3', 'ACSL4', 'ACTG1', 'ACTN1', 'ACTN4', 'ADAM12', 'ADAM9', 'ADAMTS1', 'ADAMTS2', 'ADAMTSL2', 'ADAT1', 'ADCK3', 'ADCY1', 'ADCY3', 'ADCY5', 'ADD3', 'ADM', 'ADRA2A', 'ADRA2C', 'ADRB1', 'ADRBK2', 'AFAP1L2', 'AGR2', 'AGRN', 'AHR', 'AIFM3', 'AIM1L', 'AIM1', 'AJAP1', 'AK5', 'AKAP7', 'ALDH1A1', 'ALDH1A2', 'ALDH2', 'ALDH3A2', 'ALDH3B1', 'ALDH4A1', 'ALOX5AP', 'ALPL', 'AMOTL1', 'AMOTL2', 'ANGPTL1', 'ANO6', 'ANO9', 'ANTXR2', 'AOC2', 'AP1S2', 'AP1S3', 'APOBEC2', 'APOD', 'AQP3', 'AQP4', 'ARHGAP26', 'ARHGAP29', 'ARHGAP6', 'ARHGEF17', 'ARHGEF2', 'ARHGEF3', 'ARHGEF40', 'ARHGEF6', 'ARL14', 'ARL4D', 'ARL6', 'ARMC9', 'ARNTL2', 'ARSJ', 'ARX', 'ASAP1', 'ASCL1', 'ASNS', 'ASPH', 'ASRGL1', 'ASS1', 'ASTN2', 'ASXL3', 'ATCAY', 'ATF3', 'ATF5', 'ATHL1', 'ATP10B', 'ATP11A', 'ATP1A1', 'ATP1B2', 'ATP2A3', 'ATP2B4', 'ATP8B1', 'AURKB', 'B3GNT2', 'B4GALT1', 'B4GALT5', 'BACE2', 'BACH2', 'BAIAP2L1', 'BAIAP3', 'BAMBI', 'BARX2', 'BASP1', 'BATF', 'BAZ1A', 'BBC3', 'BCHE', 'BCL2L15', 'BCL3', 'BCL6B', 'BCL6', 'BDH2', 'BHLHE41', 'BHMT2', 'BIK', 'BIRC5', 'BMF', 'BMP2', 'BMPR2', 'BNC2', 'BRIP1', 'BUB1', 'C1QL1', 'C1QTNF1', 'C1RL', 'C2CD4A', 'C2CD4B', 'CABP4', 'CACNA2D1', 'CADM2', 'CADPS2', 'CALCA', 'CALML4', 'CALU', 'CALY', 'CAMK1D', 'CAMKK1', 'CAMKK2', 'CAPN5', 'CAPN6', 'CARHSP1', 'CARTPT', 'CASP6', 'CAV2', 'CBR1', 'CCDC15', 'CCDC71L', 'CCL28', 'CCND2', 'CCNF', 'CCNG2', 'CD200', 'CD276', 'CD47', 'CD68', 'CD82', 'CDCA3', 'CDH17', 'CDH1', 'CDH23', 'CDH2', 'CDH3', 'CDHR2', 'CDK17', 'CDKN1A', 'CDKN2A', 'CDKN2B', 'CDR2', 'CDT1', 'CDX2', 'CEACAM1', 'CEBPB', 'CELF2', 'CENPF', 'CENPW', 'CERCAM', 'CFI', 'CFLAR', 'CGN', 'CHAC1', 'CHGB', 'CHN2', 'CHRM3', 'CHST1', 'CHSY1', 'CITED2', 'CITED4', 'CLCF1', 'CLCN4', 'CLDN11', 'CLDN19', 'CLDN3', 'CLDN5', 'CLDN6', 'CLDN7', 'CLEC11A', 'CLIC5', 'CLIC6', 'CLMN', 'CLMP', 'CLRN3', 'CLSPN', 'CMPK2', 'CMTM3', 'CMTM7', 'CNIH2', 'CNN2', 'CNN3', 'CNNM1', 'CNTN1', 'COCH', 'COL13A1', 'COL14A1', 'COL16A1', 'COL18A1', 'COL5A3', 'COL9A2', 'COMMD2', 'COMP', 'CORO1C', 'CORO2A', 'COX7A1', 'CPA3', 'CPLX2', 'CPM', 'CRABP2', 'CRADD', 'CREB3L1', 'CREB5', 'CREG2', 'CRIP1', 'CRLF1', 'CRTAC1', 'CRTAP', 'CTHRC1', 'CTNNA2', 'CTSB', 'CTSC', 'CTSS', 'CX3CL1', 'CXADR', 'CXCL12', 'CXCL16', 'CYB5A', 'CYBRD1', 'CYP20A1', 'CYP27B1', 'CYR61', 'CYTH3', 'DAB2IP', 'DACH1', 'DACT1', 'DAG1', 'DBN1', 'DCAF11', 'DCBLD2', 'DCUN1D2', 'DDAH2', 'DDC', 'DDIT3', 'DDX25', 'DDX51', 'DGKA', 'DHCR7', 'DHDH', 'DHODH', 'DHRS2', 'DIXDC1', 'DKK1', 'DMBT1', 'DMC1', 'DNAJB9', 'DNAJC22', 'DNER', 'DOCK4', 'DOCK5', 'DOK5', 'DPEP1', 'DPP9', 'DRAM1', 'DSC2', 'DSG2', 'DTNA', 'DUSP10', 'DUSP14', 'DUSP1', 'DUSP23', 'DUSP2', 'DUSP5', 'DZANK1', 'EBP', 'ECE1', 'ECEL1', 'EDN2', 'EFCAB7', 'EFNB1', 'EFNB3', 'EGFR', 'EGF', 'EGR1', 'EGR4', 'EHD1', 'EHD4', 'EHF', 'EIF4EBP1', 'ELAVL4', 'EMILIN1', 'EMP2', 'ENAH', 'ENG', 'ENPP2', 'EPAS1', 'EPDR1', 'EPHA2', 'EPHB2', 'EPHX1', 'EPS8L1', 'ERBB2', 'ERBB3', 'ERO1LB', 'ERO1L', 'ERRFI1', 'ESRRG', 'ETS2', 'ETV1', 'EVA1B', 'EVC2', 'EXT1', 'F10', 'F11R', 'F2RL1', 'F5', 'FA2H', 'FABP5', 'FADS1', 'FADS2', 'FAM101B', 'FAM105A', 'FAM107B', 'FAM129B', 'FAM134B', 'FAM155A', 'FAM159B', 'FAM161A', 'FAM162A', 'FAM163A', 'FAM20C', 'FAM217B', 'FAM227A', 'FAM43A', 'FAM46B', 'FAM46C', 'FAM73A', 'FAM84A', 'FAM84B', 'FBLIM1', 'FBLN1', 'FBLN5', 'FBP1', 'FBXL18', 'FBXO2', 'FBXO6', 'FCER1G', 'FDFT1', 'FERMT2', 'FEV', 'FEZ1', 'FFAR2', 'FFAR4', 'FGD2', 'FGD4', 'FGF7', 'FGFBP1', 'FGFR2', 'FGFR3', 'FGL2', 'FHL2', 'FKBP10', 'FKBP11', 'FKBP5', 'FLNA', 'FNDC4', 'FOLR1', 'FOSB', 'FOSL2', 'FOXC1', 'FOXM1', 'FOXQ1', 'FRAS1', 'FRMD4B', 'FSCN1', 'FXN', 'FXYD3', 'FXYD5', 'FXYD6', 'FZD5', 'G0S2', 'GAD2', 'GADD45A', 'GADD45B', 'GADD45G', 'GALE', 'GALM', 'GALNT2', 'GALNT3', 'GAL', 'GAS6', 'GAS7', 'GATA2', 'GCK', 'GCLC', 'GCNT1', 'GC', 'GDPD1', 'GFOD2', 'GFRA1', 'GGT1', 'GGT5', 'GHR', 'GINS3', 'GINS4', 'GJA1', 'GJA4', 'GJB1', 'GJB3', 'GJC1', 'GK5', 'GK', 'GLDC', 'GLI2', 'GLIPR2', 'GLT1D1', 'GLUL', 'GMDS', 'GMEB1', 'GMFG', 'GMNN', 'GNAL', 'GNPNAT1', 'GNPTAB', 'GOLM1', 'GOT1', 'GPC1', 'GPC6', 'GPR155', 'GPR160', 'GPR161', 'GPR4', 'GPRC5A', 'GPRC5B', 'GPSM1', 'GPT2', 'GPX3', 'GREB1', 'GSN', 'GSTM3', 'GSTM5', 'GSTP1', 'GTF2A1', 'GTPBP10', 'GUCA1B', 'GUCA2A', 'GUCY1A3', 'GULP1', 'GYLTL1B', 'H19', 'HABP2', 'HAP1', 'HBEGF', 'HCK', 'HDHD3', 'HEATR5A', 'HEBP1', 'HEG1', 'HEPH', 'HERPUD1', 'HES1', 'HIC1', 'HIF1A', 'HILPDA', 'HIP1', 'HIRA', 'HIST1H1C', 'HIST1H2BK', 'HIST2H2BE', 'HK1', 'HK2', 'HKDC1', 'HMGB3', 'HMGCS1', 'HMGN5', 'HMOX1', 'HN1', 'HNF1B', 'HNF4A', 'HNMT', 'HOMER2', 'HOPX', 'HOXB2', 'HOXB4', 'HPCAL1', 'HPGD', 'HPN', 'HS3ST1', 'HS3ST3A1', 'HS6ST2', 'HSD11B2', 'HSD17B11', 'HSPA1A', 'HSPA1B', 'HSPA5', 'HSPB6', 'HSPB8', 'HTRA1', 'HUNK', 'HYAL3', 'IBA57', 'ID1', 'ID4', 'IDH1', 'IDH2', 'IER2', 'IER3', 'IFI30', 'IFIT1', 'IFITM2', 'IFNGR1', 'IGFBP6', 'IGFBPL1', 'IGSF3', 'IKBIP', 'IKBKE', 'IKZF3', 'IL11', 'IL15RA', 'IL1RN', 'IL22RA1', 'IL33', 'IMPA2', 'INHBB', 'INPP4B', 'INSIG1', 'INSM1', 'IQGAP2', 'IRAK2', 'IRF1', 'IRS2', 'IRX1', 'IRX2', 'ISG20', 'ISL1', 'ITGA11', 'ITGA1', 'ITGA3', 'ITGA6', 'ITGAV', 'ITGB1', 'ITGB4', 'ITGB8', 'ITIH5', 'ITPR2', 'IVNS1ABP', 'IYD', 'JAM3', 'JARID2', 'JDP2', 'JMJD6', 'JPX', 'JUNB', 'JUN', 'JUP', 'KCNA5', 'KCNE3', 'KCNG1', 'KCNH6', 'KCNJ13', 'KCNJ5', 'KCNJ6', 'KCNJ8', 'KCNK16', 'KCNK5', 'KCNN2', 'KCNN3', 'KCNQ1OT1', 'KCNQ1', 'KCTD12', 'KDELC2', 'KDELR3', 'KIF11', 'KIF12', 'KIF20B', 'KIF21B', 'KIF5C', 'KIT', 'KLB', 'KLF2', 'KLF4', 'KLF5', 'KLF6', 'KLF7', 'KLHL28', 'KLHL31', 'KLHL3', 'KLK10', 'KLK11', 'KREMEN1', 'KRT15', 'KRT17', 'KRTCAP3', 'KYNU', 'L1TD1', 'L2HGDH', 'LAD1', 'LAIR1', 'LAMA1', 'LAMA3', 'LAMB1', 'LAMB2', 'LAMB3', 'LARP6', 'LASP1', 'LBH', 'LCP1', 'LDB2', 'LDLR', 'LEFTY1', 'LFNG', 'LGALS3', 'LGALS9', 'LHFPL2', 'LIFR', 'LIF', 'LIMA1', 'LIMK2', 'LIMS1', 'LIPC', 'LIPH', 'LITAF', 'LMO1', 'LMO2', 'LMO4', 'LMO7', 'LNX2', 'LOXL1', 'LOXL2', 'LPAR1', 'LRP1', 'LRP8', 'LRRC27', 'LRRC2', 'LRRC57', 'LRRC58', 'LRRIQ1', 'LRRK2', 'LRRTM2', 'LSR', 'LSS', 'LTBP4', 'LTB', 'LTF', 'LTV1', 'LURAP1L', 'LXN', 'LY6E', 'LY6H', 'LY96', 'LYN', 'LYPD1', 'LYRM7', 'LZTS1', 'MAB21L3', 'MACC1', 'MAFB', 'MAFF', 'MAL2', 'MALL', 'MAOA', 'MAOB', 'MAP3K9', 'MAPT', 'MARCKSL1', 'MAT1A', 'MCC', 'MCL1', 'MCM2', 'MCM6', 'MCOLN2', 'MDFIC', 'MDFI', 'MDGA1', 'MDK', 'MDM1', 'ME1', 'MEDAG', 'MEF2C', 'MEIS2', 'METRNL', 'METTL21A', 'MEX3A', 'MFAP2', 'MFAP4', 'MFGE8', 'MGAT4B', 'MGLL', 'MGP', 'MICAL2', 'MID1', 'MIR143HG', 'MKNK1', 'MLLT11', 'MLXIPL', 'MMACHC', 'MMP14', 'MNS1', 'MOB3B', 'MOSPD1', 'MOXD1', 'MPPED2', 'MPST', 'MPZL2', 'MRC2', 'MS4A2', 'MSMO1', 'MSRB1', 'MSRB3', 'MSX1', 'MTHFD2', 'MTUS1', 'MVP', 'MYADM', 'MYH9', 'MYL12A', 'MYL12B', 'MYL9', 'MYLK', 'MYO1C', 'MYO5B', 'MYOF', 'MYRF', 'MYZAP', 'N4BP3', 'NAA40', 'NAP1L2', 'NCAM1', 'NCF2', 'NCMAP', 'NCS1', 'NDRG1', 'NDRG2', 'NDUFA4L2', 'NDUFA4', 'NEDD9', 'NEK6', 'NEK8', 'NEURL3', 'NEUROD1', 'NEXN', 'NFATC4', 'NFKBIE', 'NFKBIZ', 'NHSL1', 'NICN1', 'NID1', 'NID2', 'NIPSNAP3B', 'NKD1', 'NLN', 'NLRC3', 'NMB', 'NOL9', 'NOTCH1', 'NOTCH3', 'NOTCH4', 'NPHS1', 'NPR1', 'NPTX2', 'NPY1R', 'NQO1', 'NR0B1', 'NR0B2', 'NR1D1', 'NR2F2', 'NR4A2', 'NR4A3', 'NR5A2', 'NR6A1', 'NRARP', 'NREP', 'NRP2', 'NSG1', 'NT5E', 'NTM', 'NTN4', 'NUAK2', 'NUGGC', 'NUSAP1', 'NYAP2', 'OAF', 'OAS3', 'OCLN', 'ODF2L', 'OGDH', 'OLFM2', 'OLFML1', 'OLFML2B', 'ORAI2', 'ORC4', 'OSGIN1', 'OSMR', 'OVOL1', 'OXR1', 'P2RX1', 'P4HA1', 'P4HA3', 'PACSIN2', 'PAG1', 'PAH', 'PALB2', 'PAMR1', 'PANK1', 'PAQR5', 'PARD6G', 'PARM1', 'PASK', 'PCDH10', 'PCDH17', 'PCDH18', 'PCDH7', 'PCK2', 'PCP4', 'PCSK2', 'PCSK5', 'PDCD4', 'PDDC1', 'PDE1A', 'PDE3B', 'PDE4A', 'PDE4C', 'PDE7B', 'PDE8B', 'PDGFA', 'PDGFB', 'PDGFC', 'PDIA4', 'PDLIM1', 'PDLIM2', 'PDLIM3', 'PDLIM4', 'PDP1', 'PDP2', 'PDX1', 'PEAK1', 'PECAM1', 'PEG10', 'PEMT', 'PGC', 'PGM1', 'PGM2L1', 'PHGDH', 'PHGR1', 'PHLDA1', 'PHLDA2', 'PHLDA3', 'PID1', 'PIEZO1', 'PIGN', 'PIK3IP1', 'PIK3R1', 'PIM1', 'PIM2', 'PIM3', 'PIWIL2', 'PKIB', 'PLA2G15', 'PLA2G4A', 'PLA2G4C', 'PLAT', 'PLBD1', 'PLCD1', 'PLCD3', 'P
#> ℹ [2026-02-11 03:38:26] Use raw method to find neighbors
#> ℹ [2026-02-11 03:38:27] Running UMAP projection
ProjectionPlot(
srt_query = srt_query,
srt_ref = srt_ref,
query_group = "celltype",
ref_group = "celltype"
)
#> Warning: No shared levels found between `names(values)` of the manual scale and the
#> data's fill values.
#> Scale for x is already present.
#> Adding another scale for x, which will replace the existing scale.
#> Scale for y is already present.
#> Adding another scale for y, which will replace the existing scale.
#> Warning: No shared levels found between `names(values)` of the manual scale and the
#> data's fill values.
#> Warning: No shared levels found between `names(values)` of the manual scale and the
#> data's fill values.
#> Warning: No shared levels found between `names(values)` of the manual scale and the
#> data's fill values.
#> Warning: Removed 6 rows containing missing values or values outside the scale range
#> (`geom_point()`).
#> Warning: Removed 6 rows containing missing values or values outside the scale range
#> (`geom_point()`).
# Projection
srt_query <- RunKNNMap(
srt_query = srt_query,
srt_ref = srt_ref,
ref_umap = "UncorrectedUMAP2D"
)
#> ℹ [2026-02-11 03:38:24] Use the features to calculate distance metric
#> ℹ [2026-02-11 03:38:24] Data type is log-normalized
#> Warning: The following features were labelled as variable in 'var.features' but had no corresponding rank in `var.features.rank` and will therefore be ignored: 'A4GALT', 'AADAC', 'AARS2', 'ABCA1', 'ABCC3', 'ABCC4', 'ABCC8', 'ABCC9', 'ABCG2', 'ABHD15', 'ABI3', 'ABL2', 'ABTB2', 'ACACB', 'ACBD7', 'ACE', 'ACHE', 'ACSL3', 'ACSL4', 'ACTG1', 'ACTN1', 'ACTN4', 'ADAM12', 'ADAM9', 'ADAMTS1', 'ADAMTS2', 'ADAMTSL2', 'ADAT1', 'ADCK3', 'ADCY1', 'ADCY3', 'ADCY5', 'ADD3', 'ADM', 'ADRA2A', 'ADRA2C', 'ADRB1', 'ADRBK2', 'AFAP1L2', 'AGR2', 'AGRN', 'AHR', 'AIFM3', 'AIM1L', 'AIM1', 'AJAP1', 'AK5', 'AKAP7', 'ALDH1A1', 'ALDH1A2', 'ALDH2', 'ALDH3A2', 'ALDH3B1', 'ALDH4A1', 'ALOX5AP', 'ALPL', 'AMOTL1', 'AMOTL2', 'ANGPTL1', 'ANO6', 'ANO9', 'ANTXR2', 'AOC2', 'AP1S2', 'AP1S3', 'APOBEC2', 'APOD', 'AQP3', 'AQP4', 'ARHGAP26', 'ARHGAP29', 'ARHGAP6', 'ARHGEF17', 'ARHGEF2', 'ARHGEF3', 'ARHGEF40', 'ARHGEF6', 'ARL14', 'ARL4D', 'ARL6', 'ARMC9', 'ARNTL2', 'ARSJ', 'ARX', 'ASAP1', 'ASCL1', 'ASNS', 'ASPH', 'ASRGL1', 'ASS1', 'ASTN2', 'ASXL3', 'ATCAY', 'ATF3', 'ATF5', 'ATHL1', 'ATP10B', 'ATP11A', 'ATP1A1', 'ATP1B2', 'ATP2A3', 'ATP2B4', 'ATP8B1', 'AURKB', 'B3GNT2', 'B4GALT1', 'B4GALT5', 'BACE2', 'BACH2', 'BAIAP2L1', 'BAIAP3', 'BAMBI', 'BARX2', 'BASP1', 'BATF', 'BAZ1A', 'BBC3', 'BCHE', 'BCL2L15', 'BCL3', 'BCL6B', 'BCL6', 'BDH2', 'BHLHE41', 'BHMT2', 'BIK', 'BIRC5', 'BMF', 'BMP2', 'BMPR2', 'BNC2', 'BRIP1', 'BUB1', 'C1QL1', 'C1QTNF1', 'C1RL', 'C2CD4A', 'C2CD4B', 'CABP4', 'CACNA2D1', 'CADM2', 'CADPS2', 'CALCA', 'CALML4', 'CALU', 'CALY', 'CAMK1D', 'CAMKK1', 'CAMKK2', 'CAPN5', 'CAPN6', 'CARHSP1', 'CARTPT', 'CASP6', 'CAV2', 'CBR1', 'CCDC15', 'CCDC71L', 'CCL28', 'CCND2', 'CCNF', 'CCNG2', 'CD200', 'CD276', 'CD47', 'CD68', 'CD82', 'CDCA3', 'CDH17', 'CDH1', 'CDH23', 'CDH2', 'CDH3', 'CDHR2', 'CDK17', 'CDKN1A', 'CDKN2A', 'CDKN2B', 'CDR2', 'CDT1', 'CDX2', 'CEACAM1', 'CEBPB', 'CELF2', 'CENPF', 'CENPW', 'CERCAM', 'CFI', 'CFLAR', 'CGN', 'CHAC1', 'CHGB', 'CHN2', 'CHRM3', 'CHST1', 'CHSY1', 'CITED2', 'CITED4', 'CLCF1', 'CLCN4', 'CLDN11', 'CLDN19', 'CLDN3', 'CLDN5', 'CLDN6', 'CLDN7', 'CLEC11A', 'CLIC5', 'CLIC6', 'CLMN', 'CLMP', 'CLRN3', 'CLSPN', 'CMPK2', 'CMTM3', 'CMTM7', 'CNIH2', 'CNN2', 'CNN3', 'CNNM1', 'CNTN1', 'COCH', 'COL13A1', 'COL14A1', 'COL16A1', 'COL18A1', 'COL5A3', 'COL9A2', 'COMMD2', 'COMP', 'CORO1C', 'CORO2A', 'COX7A1', 'CPA3', 'CPLX2', 'CPM', 'CRABP2', 'CRADD', 'CREB3L1', 'CREB5', 'CREG2', 'CRIP1', 'CRLF1', 'CRTAC1', 'CRTAP', 'CTHRC1', 'CTNNA2', 'CTSB', 'CTSC', 'CTSS', 'CX3CL1', 'CXADR', 'CXCL12', 'CXCL16', 'CYB5A', 'CYBRD1', 'CYP20A1', 'CYP27B1', 'CYR61', 'CYTH3', 'DAB2IP', 'DACH1', 'DACT1', 'DAG1', 'DBN1', 'DCAF11', 'DCBLD2', 'DCUN1D2', 'DDAH2', 'DDC', 'DDIT3', 'DDX25', 'DDX51', 'DGKA', 'DHCR7', 'DHDH', 'DHODH', 'DHRS2', 'DIXDC1', 'DKK1', 'DMBT1', 'DMC1', 'DNAJB9', 'DNAJC22', 'DNER', 'DOCK4', 'DOCK5', 'DOK5', 'DPEP1', 'DPP9', 'DRAM1', 'DSC2', 'DSG2', 'DTNA', 'DUSP10', 'DUSP14', 'DUSP1', 'DUSP23', 'DUSP2', 'DUSP5', 'DZANK1', 'EBP', 'ECE1', 'ECEL1', 'EDN2', 'EFCAB7', 'EFNB1', 'EFNB3', 'EGFR', 'EGF', 'EGR1', 'EGR4', 'EHD1', 'EHD4', 'EHF', 'EIF4EBP1', 'ELAVL4', 'EMILIN1', 'EMP2', 'ENAH', 'ENG', 'ENPP2', 'EPAS1', 'EPDR1', 'EPHA2', 'EPHB2', 'EPHX1', 'EPS8L1', 'ERBB2', 'ERBB3', 'ERO1LB', 'ERO1L', 'ERRFI1', 'ESRRG', 'ETS2', 'ETV1', 'EVA1B', 'EVC2', 'EXT1', 'F10', 'F11R', 'F2RL1', 'F5', 'FA2H', 'FABP5', 'FADS1', 'FADS2', 'FAM101B', 'FAM105A', 'FAM107B', 'FAM129B', 'FAM134B', 'FAM155A', 'FAM159B', 'FAM161A', 'FAM162A', 'FAM163A', 'FAM20C', 'FAM217B', 'FAM227A', 'FAM43A', 'FAM46B', 'FAM46C', 'FAM73A', 'FAM84A', 'FAM84B', 'FBLIM1', 'FBLN1', 'FBLN5', 'FBP1', 'FBXL18', 'FBXO2', 'FBXO6', 'FCER1G', 'FDFT1', 'FERMT2', 'FEV', 'FEZ1', 'FFAR2', 'FFAR4', 'FGD2', 'FGD4', 'FGF7', 'FGFBP1', 'FGFR2', 'FGFR3', 'FGL2', 'FHL2', 'FKBP10', 'FKBP11', 'FKBP5', 'FLNA', 'FNDC4', 'FOLR1', 'FOSB', 'FOSL2', 'FOXC1', 'FOXM1', 'FOXQ1', 'FRAS1', 'FRMD4B', 'FSCN1', 'FXN', 'FXYD3', 'FXYD5', 'FXYD6', 'FZD5', 'G0S2', 'GAD2', 'GADD45A', 'GADD45B', 'GADD45G', 'GALE', 'GALM', 'GALNT2', 'GALNT3', 'GAL', 'GAS6', 'GAS7', 'GATA2', 'GCK', 'GCLC', 'GCNT1', 'GC', 'GDPD1', 'GFOD2', 'GFRA1', 'GGT1', 'GGT5', 'GHR', 'GINS3', 'GINS4', 'GJA1', 'GJA4', 'GJB1', 'GJB3', 'GJC1', 'GK5', 'GK', 'GLDC', 'GLI2', 'GLIPR2', 'GLT1D1', 'GLUL', 'GMDS', 'GMEB1', 'GMFG', 'GMNN', 'GNAL', 'GNPNAT1', 'GNPTAB', 'GOLM1', 'GOT1', 'GPC1', 'GPC6', 'GPR155', 'GPR160', 'GPR161', 'GPR4', 'GPRC5A', 'GPRC5B', 'GPSM1', 'GPT2', 'GPX3', 'GREB1', 'GSN', 'GSTM3', 'GSTM5', 'GSTP1', 'GTF2A1', 'GTPBP10', 'GUCA1B', 'GUCA2A', 'GUCY1A3', 'GULP1', 'GYLTL1B', 'H19', 'HABP2', 'HAP1', 'HBEGF', 'HCK', 'HDHD3', 'HEATR5A', 'HEBP1', 'HEG1', 'HEPH', 'HERPUD1', 'HES1', 'HIC1', 'HIF1A', 'HILPDA', 'HIP1', 'HIRA', 'HIST1H1C', 'HIST1H2BK', 'HIST2H2BE', 'HK1', 'HK2', 'HKDC1', 'HMGB3', 'HMGCS1', 'HMGN5', 'HMOX1', 'HN1', 'HNF1B', 'HNF4A', 'HNMT', 'HOMER2', 'HOPX', 'HOXB2', 'HOXB4', 'HPCAL1', 'HPGD', 'HPN', 'HS3ST1', 'HS3ST3A1', 'HS6ST2', 'HSD11B2', 'HSD17B11', 'HSPA1A', 'HSPA1B', 'HSPA5', 'HSPB6', 'HSPB8', 'HTRA1', 'HUNK', 'HYAL3', 'IBA57', 'ID1', 'ID4', 'IDH1', 'IDH2', 'IER2', 'IER3', 'IFI30', 'IFIT1', 'IFITM2', 'IFNGR1', 'IGFBP6', 'IGFBPL1', 'IGSF3', 'IKBIP', 'IKBKE', 'IKZF3', 'IL11', 'IL15RA', 'IL1RN', 'IL22RA1', 'IL33', 'IMPA2', 'INHBB', 'INPP4B', 'INSIG1', 'INSM1', 'IQGAP2', 'IRAK2', 'IRF1', 'IRS2', 'IRX1', 'IRX2', 'ISG20', 'ISL1', 'ITGA11', 'ITGA1', 'ITGA3', 'ITGA6', 'ITGAV', 'ITGB1', 'ITGB4', 'ITGB8', 'ITIH5', 'ITPR2', 'IVNS1ABP', 'IYD', 'JAM3', 'JARID2', 'JDP2', 'JMJD6', 'JPX', 'JUNB', 'JUN', 'JUP', 'KCNA5', 'KCNE3', 'KCNG1', 'KCNH6', 'KCNJ13', 'KCNJ5', 'KCNJ6', 'KCNJ8', 'KCNK16', 'KCNK5', 'KCNN2', 'KCNN3', 'KCNQ1OT1', 'KCNQ1', 'KCTD12', 'KDELC2', 'KDELR3', 'KIF11', 'KIF12', 'KIF20B', 'KIF21B', 'KIF5C', 'KIT', 'KLB', 'KLF2', 'KLF4', 'KLF5', 'KLF6', 'KLF7', 'KLHL28', 'KLHL31', 'KLHL3', 'KLK10', 'KLK11', 'KREMEN1', 'KRT15', 'KRT17', 'KRTCAP3', 'KYNU', 'L1TD1', 'L2HGDH', 'LAD1', 'LAIR1', 'LAMA1', 'LAMA3', 'LAMB1', 'LAMB2', 'LAMB3', 'LARP6', 'LASP1', 'LBH', 'LCP1', 'LDB2', 'LDLR', 'LEFTY1', 'LFNG', 'LGALS3', 'LGALS9', 'LHFPL2', 'LIFR', 'LIF', 'LIMA1', 'LIMK2', 'LIMS1', 'LIPC', 'LIPH', 'LITAF', 'LMO1', 'LMO2', 'LMO4', 'LMO7', 'LNX2', 'LOXL1', 'LOXL2', 'LPAR1', 'LRP1', 'LRP8', 'LRRC27', 'LRRC2', 'LRRC57', 'LRRC58', 'LRRIQ1', 'LRRK2', 'LRRTM2', 'LSR', 'LSS', 'LTBP4', 'LTB', 'LTF', 'LTV1', 'LURAP1L', 'LXN', 'LY6E', 'LY6H', 'LY96', 'LYN', 'LYPD1', 'LYRM7', 'LZTS1', 'MAB21L3', 'MACC1', 'MAFB', 'MAFF', 'MAL2', 'MALL', 'MAOA', 'MAOB', 'MAP3K9', 'MAPT', 'MARCKSL1', 'MAT1A', 'MCC', 'MCL1', 'MCM2', 'MCM6', 'MCOLN2', 'MDFIC', 'MDFI', 'MDGA1', 'MDK', 'MDM1', 'ME1', 'MEDAG', 'MEF2C', 'MEIS2', 'METRNL', 'METTL21A', 'MEX3A', 'MFAP2', 'MFAP4', 'MFGE8', 'MGAT4B', 'MGLL', 'MGP', 'MICAL2', 'MID1', 'MIR143HG', 'MKNK1', 'MLLT11', 'MLXIPL', 'MMACHC', 'MMP14', 'MNS1', 'MOB3B', 'MOSPD1', 'MOXD1', 'MPPED2', 'MPST', 'MPZL2', 'MRC2', 'MS4A2', 'MSMO1', 'MSRB1', 'MSRB3', 'MSX1', 'MTHFD2', 'MTUS1', 'MVP', 'MYADM', 'MYH9', 'MYL12A', 'MYL12B', 'MYL9', 'MYLK', 'MYO1C', 'MYO5B', 'MYOF', 'MYRF', 'MYZAP', 'N4BP3', 'NAA40', 'NAP1L2', 'NCAM1', 'NCF2', 'NCMAP', 'NCS1', 'NDRG1', 'NDRG2', 'NDUFA4L2', 'NDUFA4', 'NEDD9', 'NEK6', 'NEK8', 'NEURL3', 'NEUROD1', 'NEXN', 'NFATC4', 'NFKBIE', 'NFKBIZ', 'NHSL1', 'NICN1', 'NID1', 'NID2', 'NIPSNAP3B', 'NKD1', 'NLN', 'NLRC3', 'NMB', 'NOL9', 'NOTCH1', 'NOTCH3', 'NOTCH4', 'NPHS1', 'NPR1', 'NPTX2', 'NPY1R', 'NQO1', 'NR0B1', 'NR0B2', 'NR1D1', 'NR2F2', 'NR4A2', 'NR4A3', 'NR5A2', 'NR6A1', 'NRARP', 'NREP', 'NRP2', 'NSG1', 'NT5E', 'NTM', 'NTN4', 'NUAK2', 'NUGGC', 'NUSAP1', 'NYAP2', 'OAF', 'OAS3', 'OCLN', 'ODF2L', 'OGDH', 'OLFM2', 'OLFML1', 'OLFML2B', 'ORAI2', 'ORC4', 'OSGIN1', 'OSMR', 'OVOL1', 'OXR1', 'P2RX1', 'P4HA1', 'P4HA3', 'PACSIN2', 'PAG1', 'PAH', 'PALB2', 'PAMR1', 'PANK1', 'PAQR5', 'PARD6G', 'PARM1', 'PASK', 'PCDH10', 'PCDH17', 'PCDH18', 'PCDH7', 'PCK2', 'PCP4', 'PCSK2', 'PCSK5', 'PDCD4', 'PDDC1', 'PDE1A', 'PDE3B', 'PDE4A', 'PDE4C', 'PDE7B', 'PDE8B', 'PDGFA', 'PDGFB', 'PDGFC', 'PDIA4', 'PDLIM1', 'PDLIM2', 'PDLIM3', 'PDLIM4', 'PDP1', 'PDP2', 'PDX1', 'PEAK1', 'PECAM1', 'PEG10', 'PEMT', 'PGC', 'PGM1', 'PGM2L1', 'PHGDH', 'PHGR1', 'PHLDA1', 'PHLDA2', 'PHLDA3', 'PID1', 'PIEZO1', 'PIGN', 'PIK3IP1', 'PIK3R1', 'PIM1', 'PIM2', 'PIM3', 'PIWIL2', 'PKIB', 'PLA2G15', 'PLA2G4A', 'PLA2G4C', 'PLAT', 'PLBD1', 'PLCD1', 'PLCD3', 'P
#> ℹ [2026-02-11 03:38:26] Data type is log-normalized
#> Warning: The following features were labelled as variable in 'var.features' but had no corresponding rank in `var.features.rank` and will therefore be ignored: 'A4GALT', 'AADAC', 'AARS2', 'ABCA1', 'ABCC3', 'ABCC4', 'ABCC8', 'ABCC9', 'ABCG2', 'ABHD15', 'ABI3', 'ABL2', 'ABTB2', 'ACACB', 'ACBD7', 'ACE', 'ACHE', 'ACSL3', 'ACSL4', 'ACTG1', 'ACTN1', 'ACTN4', 'ADAM12', 'ADAM9', 'ADAMTS1', 'ADAMTS2', 'ADAMTSL2', 'ADAT1', 'ADCK3', 'ADCY1', 'ADCY3', 'ADCY5', 'ADD3', 'ADM', 'ADRA2A', 'ADRA2C', 'ADRB1', 'ADRBK2', 'AFAP1L2', 'AGR2', 'AGRN', 'AHR', 'AIFM3', 'AIM1L', 'AIM1', 'AJAP1', 'AK5', 'AKAP7', 'ALDH1A1', 'ALDH1A2', 'ALDH2', 'ALDH3A2', 'ALDH3B1', 'ALDH4A1', 'ALOX5AP', 'ALPL', 'AMOTL1', 'AMOTL2', 'ANGPTL1', 'ANO6', 'ANO9', 'ANTXR2', 'AOC2', 'AP1S2', 'AP1S3', 'APOBEC2', 'APOD', 'AQP3', 'AQP4', 'ARHGAP26', 'ARHGAP29', 'ARHGAP6', 'ARHGEF17', 'ARHGEF2', 'ARHGEF3', 'ARHGEF40', 'ARHGEF6', 'ARL14', 'ARL4D', 'ARL6', 'ARMC9', 'ARNTL2', 'ARSJ', 'ARX', 'ASAP1', 'ASCL1', 'ASNS', 'ASPH', 'ASRGL1', 'ASS1', 'ASTN2', 'ASXL3', 'ATCAY', 'ATF3', 'ATF5', 'ATHL1', 'ATP10B', 'ATP11A', 'ATP1A1', 'ATP1B2', 'ATP2A3', 'ATP2B4', 'ATP8B1', 'AURKB', 'B3GNT2', 'B4GALT1', 'B4GALT5', 'BACE2', 'BACH2', 'BAIAP2L1', 'BAIAP3', 'BAMBI', 'BARX2', 'BASP1', 'BATF', 'BAZ1A', 'BBC3', 'BCHE', 'BCL2L15', 'BCL3', 'BCL6B', 'BCL6', 'BDH2', 'BHLHE41', 'BHMT2', 'BIK', 'BIRC5', 'BMF', 'BMP2', 'BMPR2', 'BNC2', 'BRIP1', 'BUB1', 'C1QL1', 'C1QTNF1', 'C1RL', 'C2CD4A', 'C2CD4B', 'CABP4', 'CACNA2D1', 'CADM2', 'CADPS2', 'CALCA', 'CALML4', 'CALU', 'CALY', 'CAMK1D', 'CAMKK1', 'CAMKK2', 'CAPN5', 'CAPN6', 'CARHSP1', 'CARTPT', 'CASP6', 'CAV2', 'CBR1', 'CCDC15', 'CCDC71L', 'CCL28', 'CCND2', 'CCNF', 'CCNG2', 'CD200', 'CD276', 'CD47', 'CD68', 'CD82', 'CDCA3', 'CDH17', 'CDH1', 'CDH23', 'CDH2', 'CDH3', 'CDHR2', 'CDK17', 'CDKN1A', 'CDKN2A', 'CDKN2B', 'CDR2', 'CDT1', 'CDX2', 'CEACAM1', 'CEBPB', 'CELF2', 'CENPF', 'CENPW', 'CERCAM', 'CFI', 'CFLAR', 'CGN', 'CHAC1', 'CHGB', 'CHN2', 'CHRM3', 'CHST1', 'CHSY1', 'CITED2', 'CITED4', 'CLCF1', 'CLCN4', 'CLDN11', 'CLDN19', 'CLDN3', 'CLDN5', 'CLDN6', 'CLDN7', 'CLEC11A', 'CLIC5', 'CLIC6', 'CLMN', 'CLMP', 'CLRN3', 'CLSPN', 'CMPK2', 'CMTM3', 'CMTM7', 'CNIH2', 'CNN2', 'CNN3', 'CNNM1', 'CNTN1', 'COCH', 'COL13A1', 'COL14A1', 'COL16A1', 'COL18A1', 'COL5A3', 'COL9A2', 'COMMD2', 'COMP', 'CORO1C', 'CORO2A', 'COX7A1', 'CPA3', 'CPLX2', 'CPM', 'CRABP2', 'CRADD', 'CREB3L1', 'CREB5', 'CREG2', 'CRIP1', 'CRLF1', 'CRTAC1', 'CRTAP', 'CTHRC1', 'CTNNA2', 'CTSB', 'CTSC', 'CTSS', 'CX3CL1', 'CXADR', 'CXCL12', 'CXCL16', 'CYB5A', 'CYBRD1', 'CYP20A1', 'CYP27B1', 'CYR61', 'CYTH3', 'DAB2IP', 'DACH1', 'DACT1', 'DAG1', 'DBN1', 'DCAF11', 'DCBLD2', 'DCUN1D2', 'DDAH2', 'DDC', 'DDIT3', 'DDX25', 'DDX51', 'DGKA', 'DHCR7', 'DHDH', 'DHODH', 'DHRS2', 'DIXDC1', 'DKK1', 'DMBT1', 'DMC1', 'DNAJB9', 'DNAJC22', 'DNER', 'DOCK4', 'DOCK5', 'DOK5', 'DPEP1', 'DPP9', 'DRAM1', 'DSC2', 'DSG2', 'DTNA', 'DUSP10', 'DUSP14', 'DUSP1', 'DUSP23', 'DUSP2', 'DUSP5', 'DZANK1', 'EBP', 'ECE1', 'ECEL1', 'EDN2', 'EFCAB7', 'EFNB1', 'EFNB3', 'EGFR', 'EGF', 'EGR1', 'EGR4', 'EHD1', 'EHD4', 'EHF', 'EIF4EBP1', 'ELAVL4', 'EMILIN1', 'EMP2', 'ENAH', 'ENG', 'ENPP2', 'EPAS1', 'EPDR1', 'EPHA2', 'EPHB2', 'EPHX1', 'EPS8L1', 'ERBB2', 'ERBB3', 'ERO1LB', 'ERO1L', 'ERRFI1', 'ESRRG', 'ETS2', 'ETV1', 'EVA1B', 'EVC2', 'EXT1', 'F10', 'F11R', 'F2RL1', 'F5', 'FA2H', 'FABP5', 'FADS1', 'FADS2', 'FAM101B', 'FAM105A', 'FAM107B', 'FAM129B', 'FAM134B', 'FAM155A', 'FAM159B', 'FAM161A', 'FAM162A', 'FAM163A', 'FAM20C', 'FAM217B', 'FAM227A', 'FAM43A', 'FAM46B', 'FAM46C', 'FAM73A', 'FAM84A', 'FAM84B', 'FBLIM1', 'FBLN1', 'FBLN5', 'FBP1', 'FBXL18', 'FBXO2', 'FBXO6', 'FCER1G', 'FDFT1', 'FERMT2', 'FEV', 'FEZ1', 'FFAR2', 'FFAR4', 'FGD2', 'FGD4', 'FGF7', 'FGFBP1', 'FGFR2', 'FGFR3', 'FGL2', 'FHL2', 'FKBP10', 'FKBP11', 'FKBP5', 'FLNA', 'FNDC4', 'FOLR1', 'FOSB', 'FOSL2', 'FOXC1', 'FOXM1', 'FOXQ1', 'FRAS1', 'FRMD4B', 'FSCN1', 'FXN', 'FXYD3', 'FXYD5', 'FXYD6', 'FZD5', 'G0S2', 'GAD2', 'GADD45A', 'GADD45B', 'GADD45G', 'GALE', 'GALM', 'GALNT2', 'GALNT3', 'GAL', 'GAS6', 'GAS7', 'GATA2', 'GCK', 'GCLC', 'GCNT1', 'GC', 'GDPD1', 'GFOD2', 'GFRA1', 'GGT1', 'GGT5', 'GHR', 'GINS3', 'GINS4', 'GJA1', 'GJA4', 'GJB1', 'GJB3', 'GJC1', 'GK5', 'GK', 'GLDC', 'GLI2', 'GLIPR2', 'GLT1D1', 'GLUL', 'GMDS', 'GMEB1', 'GMFG', 'GMNN', 'GNAL', 'GNPNAT1', 'GNPTAB', 'GOLM1', 'GOT1', 'GPC1', 'GPC6', 'GPR155', 'GPR160', 'GPR161', 'GPR4', 'GPRC5A', 'GPRC5B', 'GPSM1', 'GPT2', 'GPX3', 'GREB1', 'GSN', 'GSTM3', 'GSTM5', 'GSTP1', 'GTF2A1', 'GTPBP10', 'GUCA1B', 'GUCA2A', 'GUCY1A3', 'GULP1', 'GYLTL1B', 'H19', 'HABP2', 'HAP1', 'HBEGF', 'HCK', 'HDHD3', 'HEATR5A', 'HEBP1', 'HEG1', 'HEPH', 'HERPUD1', 'HES1', 'HIC1', 'HIF1A', 'HILPDA', 'HIP1', 'HIRA', 'HIST1H1C', 'HIST1H2BK', 'HIST2H2BE', 'HK1', 'HK2', 'HKDC1', 'HMGB3', 'HMGCS1', 'HMGN5', 'HMOX1', 'HN1', 'HNF1B', 'HNF4A', 'HNMT', 'HOMER2', 'HOPX', 'HOXB2', 'HOXB4', 'HPCAL1', 'HPGD', 'HPN', 'HS3ST1', 'HS3ST3A1', 'HS6ST2', 'HSD11B2', 'HSD17B11', 'HSPA1A', 'HSPA1B', 'HSPA5', 'HSPB6', 'HSPB8', 'HTRA1', 'HUNK', 'HYAL3', 'IBA57', 'ID1', 'ID4', 'IDH1', 'IDH2', 'IER2', 'IER3', 'IFI30', 'IFIT1', 'IFITM2', 'IFNGR1', 'IGFBP6', 'IGFBPL1', 'IGSF3', 'IKBIP', 'IKBKE', 'IKZF3', 'IL11', 'IL15RA', 'IL1RN', 'IL22RA1', 'IL33', 'IMPA2', 'INHBB', 'INPP4B', 'INSIG1', 'INSM1', 'IQGAP2', 'IRAK2', 'IRF1', 'IRS2', 'IRX1', 'IRX2', 'ISG20', 'ISL1', 'ITGA11', 'ITGA1', 'ITGA3', 'ITGA6', 'ITGAV', 'ITGB1', 'ITGB4', 'ITGB8', 'ITIH5', 'ITPR2', 'IVNS1ABP', 'IYD', 'JAM3', 'JARID2', 'JDP2', 'JMJD6', 'JPX', 'JUNB', 'JUN', 'JUP', 'KCNA5', 'KCNE3', 'KCNG1', 'KCNH6', 'KCNJ13', 'KCNJ5', 'KCNJ6', 'KCNJ8', 'KCNK16', 'KCNK5', 'KCNN2', 'KCNN3', 'KCNQ1OT1', 'KCNQ1', 'KCTD12', 'KDELC2', 'KDELR3', 'KIF11', 'KIF12', 'KIF20B', 'KIF21B', 'KIF5C', 'KIT', 'KLB', 'KLF2', 'KLF4', 'KLF5', 'KLF6', 'KLF7', 'KLHL28', 'KLHL31', 'KLHL3', 'KLK10', 'KLK11', 'KREMEN1', 'KRT15', 'KRT17', 'KRTCAP3', 'KYNU', 'L1TD1', 'L2HGDH', 'LAD1', 'LAIR1', 'LAMA1', 'LAMA3', 'LAMB1', 'LAMB2', 'LAMB3', 'LARP6', 'LASP1', 'LBH', 'LCP1', 'LDB2', 'LDLR', 'LEFTY1', 'LFNG', 'LGALS3', 'LGALS9', 'LHFPL2', 'LIFR', 'LIF', 'LIMA1', 'LIMK2', 'LIMS1', 'LIPC', 'LIPH', 'LITAF', 'LMO1', 'LMO2', 'LMO4', 'LMO7', 'LNX2', 'LOXL1', 'LOXL2', 'LPAR1', 'LRP1', 'LRP8', 'LRRC27', 'LRRC2', 'LRRC57', 'LRRC58', 'LRRIQ1', 'LRRK2', 'LRRTM2', 'LSR', 'LSS', 'LTBP4', 'LTB', 'LTF', 'LTV1', 'LURAP1L', 'LXN', 'LY6E', 'LY6H', 'LY96', 'LYN', 'LYPD1', 'LYRM7', 'LZTS1', 'MAB21L3', 'MACC1', 'MAFB', 'MAFF', 'MAL2', 'MALL', 'MAOA', 'MAOB', 'MAP3K9', 'MAPT', 'MARCKSL1', 'MAT1A', 'MCC', 'MCL1', 'MCM2', 'MCM6', 'MCOLN2', 'MDFIC', 'MDFI', 'MDGA1', 'MDK', 'MDM1', 'ME1', 'MEDAG', 'MEF2C', 'MEIS2', 'METRNL', 'METTL21A', 'MEX3A', 'MFAP2', 'MFAP4', 'MFGE8', 'MGAT4B', 'MGLL', 'MGP', 'MICAL2', 'MID1', 'MIR143HG', 'MKNK1', 'MLLT11', 'MLXIPL', 'MMACHC', 'MMP14', 'MNS1', 'MOB3B', 'MOSPD1', 'MOXD1', 'MPPED2', 'MPST', 'MPZL2', 'MRC2', 'MS4A2', 'MSMO1', 'MSRB1', 'MSRB3', 'MSX1', 'MTHFD2', 'MTUS1', 'MVP', 'MYADM', 'MYH9', 'MYL12A', 'MYL12B', 'MYL9', 'MYLK', 'MYO1C', 'MYO5B', 'MYOF', 'MYRF', 'MYZAP', 'N4BP3', 'NAA40', 'NAP1L2', 'NCAM1', 'NCF2', 'NCMAP', 'NCS1', 'NDRG1', 'NDRG2', 'NDUFA4L2', 'NDUFA4', 'NEDD9', 'NEK6', 'NEK8', 'NEURL3', 'NEUROD1', 'NEXN', 'NFATC4', 'NFKBIE', 'NFKBIZ', 'NHSL1', 'NICN1', 'NID1', 'NID2', 'NIPSNAP3B', 'NKD1', 'NLN', 'NLRC3', 'NMB', 'NOL9', 'NOTCH1', 'NOTCH3', 'NOTCH4', 'NPHS1', 'NPR1', 'NPTX2', 'NPY1R', 'NQO1', 'NR0B1', 'NR0B2', 'NR1D1', 'NR2F2', 'NR4A2', 'NR4A3', 'NR5A2', 'NR6A1', 'NRARP', 'NREP', 'NRP2', 'NSG1', 'NT5E', 'NTM', 'NTN4', 'NUAK2', 'NUGGC', 'NUSAP1', 'NYAP2', 'OAF', 'OAS3', 'OCLN', 'ODF2L', 'OGDH', 'OLFM2', 'OLFML1', 'OLFML2B', 'ORAI2', 'ORC4', 'OSGIN1', 'OSMR', 'OVOL1', 'OXR1', 'P2RX1', 'P4HA1', 'P4HA3', 'PACSIN2', 'PAG1', 'PAH', 'PALB2', 'PAMR1', 'PANK1', 'PAQR5', 'PARD6G', 'PARM1', 'PASK', 'PCDH10', 'PCDH17', 'PCDH18', 'PCDH7', 'PCK2', 'PCP4', 'PCSK2', 'PCSK5', 'PDCD4', 'PDDC1', 'PDE1A', 'PDE3B', 'PDE4A', 'PDE4C', 'PDE7B', 'PDE8B', 'PDGFA', 'PDGFB', 'PDGFC', 'PDIA4', 'PDLIM1', 'PDLIM2', 'PDLIM3', 'PDLIM4', 'PDP1', 'PDP2', 'PDX1', 'PEAK1', 'PECAM1', 'PEG10', 'PEMT', 'PGC', 'PGM1', 'PGM2L1', 'PHGDH', 'PHGR1', 'PHLDA1', 'PHLDA2', 'PHLDA3', 'PID1', 'PIEZO1', 'PIGN', 'PIK3IP1', 'PIK3R1', 'PIM1', 'PIM2', 'PIM3', 'PIWIL2', 'PKIB', 'PLA2G15', 'PLA2G4A', 'PLA2G4C', 'PLAT', 'PLBD1', 'PLCD1', 'PLCD3', 'P
#> ℹ [2026-02-11 03:38:26] Use 2000 features to calculate distance
#> Warning: The following features were labelled as variable in 'var.features' but had no corresponding rank in `var.features.rank` and will therefore be ignored: 'A4GALT', 'AADAC', 'AARS2', 'ABCA1', 'ABCC3', 'ABCC4', 'ABCC8', 'ABCC9', 'ABCG2', 'ABHD15', 'ABI3', 'ABL2', 'ABTB2', 'ACACB', 'ACBD7', 'ACE', 'ACHE', 'ACSL3', 'ACSL4', 'ACTG1', 'ACTN1', 'ACTN4', 'ADAM12', 'ADAM9', 'ADAMTS1', 'ADAMTS2', 'ADAMTSL2', 'ADAT1', 'ADCK3', 'ADCY1', 'ADCY3', 'ADCY5', 'ADD3', 'ADM', 'ADRA2A', 'ADRA2C', 'ADRB1', 'ADRBK2', 'AFAP1L2', 'AGR2', 'AGRN', 'AHR', 'AIFM3', 'AIM1L', 'AIM1', 'AJAP1', 'AK5', 'AKAP7', 'ALDH1A1', 'ALDH1A2', 'ALDH2', 'ALDH3A2', 'ALDH3B1', 'ALDH4A1', 'ALOX5AP', 'ALPL', 'AMOTL1', 'AMOTL2', 'ANGPTL1', 'ANO6', 'ANO9', 'ANTXR2', 'AOC2', 'AP1S2', 'AP1S3', 'APOBEC2', 'APOD', 'AQP3', 'AQP4', 'ARHGAP26', 'ARHGAP29', 'ARHGAP6', 'ARHGEF17', 'ARHGEF2', 'ARHGEF3', 'ARHGEF40', 'ARHGEF6', 'ARL14', 'ARL4D', 'ARL6', 'ARMC9', 'ARNTL2', 'ARSJ', 'ARX', 'ASAP1', 'ASCL1', 'ASNS', 'ASPH', 'ASRGL1', 'ASS1', 'ASTN2', 'ASXL3', 'ATCAY', 'ATF3', 'ATF5', 'ATHL1', 'ATP10B', 'ATP11A', 'ATP1A1', 'ATP1B2', 'ATP2A3', 'ATP2B4', 'ATP8B1', 'AURKB', 'B3GNT2', 'B4GALT1', 'B4GALT5', 'BACE2', 'BACH2', 'BAIAP2L1', 'BAIAP3', 'BAMBI', 'BARX2', 'BASP1', 'BATF', 'BAZ1A', 'BBC3', 'BCHE', 'BCL2L15', 'BCL3', 'BCL6B', 'BCL6', 'BDH2', 'BHLHE41', 'BHMT2', 'BIK', 'BIRC5', 'BMF', 'BMP2', 'BMPR2', 'BNC2', 'BRIP1', 'BUB1', 'C1QL1', 'C1QTNF1', 'C1RL', 'C2CD4A', 'C2CD4B', 'CABP4', 'CACNA2D1', 'CADM2', 'CADPS2', 'CALCA', 'CALML4', 'CALU', 'CALY', 'CAMK1D', 'CAMKK1', 'CAMKK2', 'CAPN5', 'CAPN6', 'CARHSP1', 'CARTPT', 'CASP6', 'CAV2', 'CBR1', 'CCDC15', 'CCDC71L', 'CCL28', 'CCND2', 'CCNF', 'CCNG2', 'CD200', 'CD276', 'CD47', 'CD68', 'CD82', 'CDCA3', 'CDH17', 'CDH1', 'CDH23', 'CDH2', 'CDH3', 'CDHR2', 'CDK17', 'CDKN1A', 'CDKN2A', 'CDKN2B', 'CDR2', 'CDT1', 'CDX2', 'CEACAM1', 'CEBPB', 'CELF2', 'CENPF', 'CENPW', 'CERCAM', 'CFI', 'CFLAR', 'CGN', 'CHAC1', 'CHGB', 'CHN2', 'CHRM3', 'CHST1', 'CHSY1', 'CITED2', 'CITED4', 'CLCF1', 'CLCN4', 'CLDN11', 'CLDN19', 'CLDN3', 'CLDN5', 'CLDN6', 'CLDN7', 'CLEC11A', 'CLIC5', 'CLIC6', 'CLMN', 'CLMP', 'CLRN3', 'CLSPN', 'CMPK2', 'CMTM3', 'CMTM7', 'CNIH2', 'CNN2', 'CNN3', 'CNNM1', 'CNTN1', 'COCH', 'COL13A1', 'COL14A1', 'COL16A1', 'COL18A1', 'COL5A3', 'COL9A2', 'COMMD2', 'COMP', 'CORO1C', 'CORO2A', 'COX7A1', 'CPA3', 'CPLX2', 'CPM', 'CRABP2', 'CRADD', 'CREB3L1', 'CREB5', 'CREG2', 'CRIP1', 'CRLF1', 'CRTAC1', 'CRTAP', 'CTHRC1', 'CTNNA2', 'CTSB', 'CTSC', 'CTSS', 'CX3CL1', 'CXADR', 'CXCL12', 'CXCL16', 'CYB5A', 'CYBRD1', 'CYP20A1', 'CYP27B1', 'CYR61', 'CYTH3', 'DAB2IP', 'DACH1', 'DACT1', 'DAG1', 'DBN1', 'DCAF11', 'DCBLD2', 'DCUN1D2', 'DDAH2', 'DDC', 'DDIT3', 'DDX25', 'DDX51', 'DGKA', 'DHCR7', 'DHDH', 'DHODH', 'DHRS2', 'DIXDC1', 'DKK1', 'DMBT1', 'DMC1', 'DNAJB9', 'DNAJC22', 'DNER', 'DOCK4', 'DOCK5', 'DOK5', 'DPEP1', 'DPP9', 'DRAM1', 'DSC2', 'DSG2', 'DTNA', 'DUSP10', 'DUSP14', 'DUSP1', 'DUSP23', 'DUSP2', 'DUSP5', 'DZANK1', 'EBP', 'ECE1', 'ECEL1', 'EDN2', 'EFCAB7', 'EFNB1', 'EFNB3', 'EGFR', 'EGF', 'EGR1', 'EGR4', 'EHD1', 'EHD4', 'EHF', 'EIF4EBP1', 'ELAVL4', 'EMILIN1', 'EMP2', 'ENAH', 'ENG', 'ENPP2', 'EPAS1', 'EPDR1', 'EPHA2', 'EPHB2', 'EPHX1', 'EPS8L1', 'ERBB2', 'ERBB3', 'ERO1LB', 'ERO1L', 'ERRFI1', 'ESRRG', 'ETS2', 'ETV1', 'EVA1B', 'EVC2', 'EXT1', 'F10', 'F11R', 'F2RL1', 'F5', 'FA2H', 'FABP5', 'FADS1', 'FADS2', 'FAM101B', 'FAM105A', 'FAM107B', 'FAM129B', 'FAM134B', 'FAM155A', 'FAM159B', 'FAM161A', 'FAM162A', 'FAM163A', 'FAM20C', 'FAM217B', 'FAM227A', 'FAM43A', 'FAM46B', 'FAM46C', 'FAM73A', 'FAM84A', 'FAM84B', 'FBLIM1', 'FBLN1', 'FBLN5', 'FBP1', 'FBXL18', 'FBXO2', 'FBXO6', 'FCER1G', 'FDFT1', 'FERMT2', 'FEV', 'FEZ1', 'FFAR2', 'FFAR4', 'FGD2', 'FGD4', 'FGF7', 'FGFBP1', 'FGFR2', 'FGFR3', 'FGL2', 'FHL2', 'FKBP10', 'FKBP11', 'FKBP5', 'FLNA', 'FNDC4', 'FOLR1', 'FOSB', 'FOSL2', 'FOXC1', 'FOXM1', 'FOXQ1', 'FRAS1', 'FRMD4B', 'FSCN1', 'FXN', 'FXYD3', 'FXYD5', 'FXYD6', 'FZD5', 'G0S2', 'GAD2', 'GADD45A', 'GADD45B', 'GADD45G', 'GALE', 'GALM', 'GALNT2', 'GALNT3', 'GAL', 'GAS6', 'GAS7', 'GATA2', 'GCK', 'GCLC', 'GCNT1', 'GC', 'GDPD1', 'GFOD2', 'GFRA1', 'GGT1', 'GGT5', 'GHR', 'GINS3', 'GINS4', 'GJA1', 'GJA4', 'GJB1', 'GJB3', 'GJC1', 'GK5', 'GK', 'GLDC', 'GLI2', 'GLIPR2', 'GLT1D1', 'GLUL', 'GMDS', 'GMEB1', 'GMFG', 'GMNN', 'GNAL', 'GNPNAT1', 'GNPTAB', 'GOLM1', 'GOT1', 'GPC1', 'GPC6', 'GPR155', 'GPR160', 'GPR161', 'GPR4', 'GPRC5A', 'GPRC5B', 'GPSM1', 'GPT2', 'GPX3', 'GREB1', 'GSN', 'GSTM3', 'GSTM5', 'GSTP1', 'GTF2A1', 'GTPBP10', 'GUCA1B', 'GUCA2A', 'GUCY1A3', 'GULP1', 'GYLTL1B', 'H19', 'HABP2', 'HAP1', 'HBEGF', 'HCK', 'HDHD3', 'HEATR5A', 'HEBP1', 'HEG1', 'HEPH', 'HERPUD1', 'HES1', 'HIC1', 'HIF1A', 'HILPDA', 'HIP1', 'HIRA', 'HIST1H1C', 'HIST1H2BK', 'HIST2H2BE', 'HK1', 'HK2', 'HKDC1', 'HMGB3', 'HMGCS1', 'HMGN5', 'HMOX1', 'HN1', 'HNF1B', 'HNF4A', 'HNMT', 'HOMER2', 'HOPX', 'HOXB2', 'HOXB4', 'HPCAL1', 'HPGD', 'HPN', 'HS3ST1', 'HS3ST3A1', 'HS6ST2', 'HSD11B2', 'HSD17B11', 'HSPA1A', 'HSPA1B', 'HSPA5', 'HSPB6', 'HSPB8', 'HTRA1', 'HUNK', 'HYAL3', 'IBA57', 'ID1', 'ID4', 'IDH1', 'IDH2', 'IER2', 'IER3', 'IFI30', 'IFIT1', 'IFITM2', 'IFNGR1', 'IGFBP6', 'IGFBPL1', 'IGSF3', 'IKBIP', 'IKBKE', 'IKZF3', 'IL11', 'IL15RA', 'IL1RN', 'IL22RA1', 'IL33', 'IMPA2', 'INHBB', 'INPP4B', 'INSIG1', 'INSM1', 'IQGAP2', 'IRAK2', 'IRF1', 'IRS2', 'IRX1', 'IRX2', 'ISG20', 'ISL1', 'ITGA11', 'ITGA1', 'ITGA3', 'ITGA6', 'ITGAV', 'ITGB1', 'ITGB4', 'ITGB8', 'ITIH5', 'ITPR2', 'IVNS1ABP', 'IYD', 'JAM3', 'JARID2', 'JDP2', 'JMJD6', 'JPX', 'JUNB', 'JUN', 'JUP', 'KCNA5', 'KCNE3', 'KCNG1', 'KCNH6', 'KCNJ13', 'KCNJ5', 'KCNJ6', 'KCNJ8', 'KCNK16', 'KCNK5', 'KCNN2', 'KCNN3', 'KCNQ1OT1', 'KCNQ1', 'KCTD12', 'KDELC2', 'KDELR3', 'KIF11', 'KIF12', 'KIF20B', 'KIF21B', 'KIF5C', 'KIT', 'KLB', 'KLF2', 'KLF4', 'KLF5', 'KLF6', 'KLF7', 'KLHL28', 'KLHL31', 'KLHL3', 'KLK10', 'KLK11', 'KREMEN1', 'KRT15', 'KRT17', 'KRTCAP3', 'KYNU', 'L1TD1', 'L2HGDH', 'LAD1', 'LAIR1', 'LAMA1', 'LAMA3', 'LAMB1', 'LAMB2', 'LAMB3', 'LARP6', 'LASP1', 'LBH', 'LCP1', 'LDB2', 'LDLR', 'LEFTY1', 'LFNG', 'LGALS3', 'LGALS9', 'LHFPL2', 'LIFR', 'LIF', 'LIMA1', 'LIMK2', 'LIMS1', 'LIPC', 'LIPH', 'LITAF', 'LMO1', 'LMO2', 'LMO4', 'LMO7', 'LNX2', 'LOXL1', 'LOXL2', 'LPAR1', 'LRP1', 'LRP8', 'LRRC27', 'LRRC2', 'LRRC57', 'LRRC58', 'LRRIQ1', 'LRRK2', 'LRRTM2', 'LSR', 'LSS', 'LTBP4', 'LTB', 'LTF', 'LTV1', 'LURAP1L', 'LXN', 'LY6E', 'LY6H', 'LY96', 'LYN', 'LYPD1', 'LYRM7', 'LZTS1', 'MAB21L3', 'MACC1', 'MAFB', 'MAFF', 'MAL2', 'MALL', 'MAOA', 'MAOB', 'MAP3K9', 'MAPT', 'MARCKSL1', 'MAT1A', 'MCC', 'MCL1', 'MCM2', 'MCM6', 'MCOLN2', 'MDFIC', 'MDFI', 'MDGA1', 'MDK', 'MDM1', 'ME1', 'MEDAG', 'MEF2C', 'MEIS2', 'METRNL', 'METTL21A', 'MEX3A', 'MFAP2', 'MFAP4', 'MFGE8', 'MGAT4B', 'MGLL', 'MGP', 'MICAL2', 'MID1', 'MIR143HG', 'MKNK1', 'MLLT11', 'MLXIPL', 'MMACHC', 'MMP14', 'MNS1', 'MOB3B', 'MOSPD1', 'MOXD1', 'MPPED2', 'MPST', 'MPZL2', 'MRC2', 'MS4A2', 'MSMO1', 'MSRB1', 'MSRB3', 'MSX1', 'MTHFD2', 'MTUS1', 'MVP', 'MYADM', 'MYH9', 'MYL12A', 'MYL12B', 'MYL9', 'MYLK', 'MYO1C', 'MYO5B', 'MYOF', 'MYRF', 'MYZAP', 'N4BP3', 'NAA40', 'NAP1L2', 'NCAM1', 'NCF2', 'NCMAP', 'NCS1', 'NDRG1', 'NDRG2', 'NDUFA4L2', 'NDUFA4', 'NEDD9', 'NEK6', 'NEK8', 'NEURL3', 'NEUROD1', 'NEXN', 'NFATC4', 'NFKBIE', 'NFKBIZ', 'NHSL1', 'NICN1', 'NID1', 'NID2', 'NIPSNAP3B', 'NKD1', 'NLN', 'NLRC3', 'NMB', 'NOL9', 'NOTCH1', 'NOTCH3', 'NOTCH4', 'NPHS1', 'NPR1', 'NPTX2', 'NPY1R', 'NQO1', 'NR0B1', 'NR0B2', 'NR1D1', 'NR2F2', 'NR4A2', 'NR4A3', 'NR5A2', 'NR6A1', 'NRARP', 'NREP', 'NRP2', 'NSG1', 'NT5E', 'NTM', 'NTN4', 'NUAK2', 'NUGGC', 'NUSAP1', 'NYAP2', 'OAF', 'OAS3', 'OCLN', 'ODF2L', 'OGDH', 'OLFM2', 'OLFML1', 'OLFML2B', 'ORAI2', 'ORC4', 'OSGIN1', 'OSMR', 'OVOL1', 'OXR1', 'P2RX1', 'P4HA1', 'P4HA3', 'PACSIN2', 'PAG1', 'PAH', 'PALB2', 'PAMR1', 'PANK1', 'PAQR5', 'PARD6G', 'PARM1', 'PASK', 'PCDH10', 'PCDH17', 'PCDH18', 'PCDH7', 'PCK2', 'PCP4', 'PCSK2', 'PCSK5', 'PDCD4', 'PDDC1', 'PDE1A', 'PDE3B', 'PDE4A', 'PDE4C', 'PDE7B', 'PDE8B', 'PDGFA', 'PDGFB', 'PDGFC', 'PDIA4', 'PDLIM1', 'PDLIM2', 'PDLIM3', 'PDLIM4', 'PDP1', 'PDP2', 'PDX1', 'PEAK1', 'PECAM1', 'PEG10', 'PEMT', 'PGC', 'PGM1', 'PGM2L1', 'PHGDH', 'PHGR1', 'PHLDA1', 'PHLDA2', 'PHLDA3', 'PID1', 'PIEZO1', 'PIGN', 'PIK3IP1', 'PIK3R1', 'PIM1', 'PIM2', 'PIM3', 'PIWIL2', 'PKIB', 'PLA2G15', 'PLA2G4A', 'PLA2G4C', 'PLAT', 'PLBD1', 'PLCD1', 'PLCD3', 'P
#> ℹ [2026-02-11 03:38:26] Use raw method to find neighbors
#> ℹ [2026-02-11 03:38:27] Running UMAP projection
ProjectionPlot(
srt_query = srt_query,
srt_ref = srt_ref,
query_group = "celltype",
ref_group = "celltype"
)
#> Warning: No shared levels found between `names(values)` of the manual scale and the
#> data's fill values.
#> Scale for x is already present.
#> Adding another scale for x, which will replace the existing scale.
#> Scale for y is already present.
#> Adding another scale for y, which will replace the existing scale.
#> Warning: No shared levels found between `names(values)` of the manual scale and the
#> data's fill values.
#> Warning: No shared levels found between `names(values)` of the manual scale and the
#> data's fill values.
#> Warning: No shared levels found between `names(values)` of the manual scale and the
#> data's fill values.
#> Warning: Removed 6 rows containing missing values or values outside the scale range
#> (`geom_point()`).
#> Warning: Removed 6 rows containing missing values or values outside the scale range
#> (`geom_point()`).