Calculates dynamic features for lineages
Usage
RunDynamicFeatures(
srt,
lineages,
features = NULL,
suffix = lineages,
n_candidates = 1000,
minfreq = 5,
family = NULL,
layer = "counts",
assay = NULL,
libsize = NULL,
cores = 1,
verbose = TRUE,
seed = 11
)Arguments
- srt
A Seurat object.
- lineages
A character vector specifying the lineage names for which dynamic features should be calculated.
- features
A character vector of features to use. If
NULL, n_candidates must be provided.- suffix
A character vector specifying the suffix to append to the output layer names for each lineage. Default is the lineage names.
- n_candidates
A number of candidate features to select when features is
NULL. Default is1000.- minfreq
An integer specifying the minimum frequency threshold for candidate features. Features with a frequency less than minfreq will be excluded. Default is
5.- family
A character or character vector specifying the family of distributions to use for the generalized additive models (GAMs). If family is set to NULL, the appropriate family will be automatically determined based on the data. If length(family) is 1, the same family will be used for all features. Otherwise, family must have the same length as features.
- layer
Which layer to use. Default is
"counts".- assay
Which assay to use. If
NULL, the default assay of the Seurat object will be used.- libsize
A numeric or numeric vector specifying the library size correction factors for each cell. If NULL, the library size correction factors will be calculated based on the expression matrix. If length(libsize) is 1, the same value will be used for all cells. Otherwise, libsize must have the same length as the number of cells in srt. Default is
NULL.- cores
The number of cores to use for parallelization with foreach::foreach. Default is
1.- verbose
Whether to print the message. Default is
TRUE.- seed
Random seed for reproducibility. Default is
11.
Value
Returns the modified Seurat object with the calculated dynamic features stored in the tools slot.
Examples
data(pancreas_sub)
pancreas_sub <- standard_scop(pancreas_sub)
#> ℹ [2026-02-11 03:58:09] Start standard scop workflow...
#> ℹ [2026-02-11 03:58:10] Checking a list of <Seurat>...
#> ! [2026-02-11 03:58:10] Data 1/1 of the `srt_list` is "unknown"
#> ℹ [2026-02-11 03:58:10] Perform `NormalizeData()` with `normalization.method = 'LogNormalize'` on the data 1/1 of the `srt_list`...
#> ℹ [2026-02-11 03:58:12] Perform `Seurat::FindVariableFeatures()` on the data 1/1 of the `srt_list`...
#> ℹ [2026-02-11 03:58:13] Use the separate HVF from srt_list
#> ℹ [2026-02-11 03:58:13] Number of available HVF: 2000
#> ℹ [2026-02-11 03:58:13] Finished check
#> ℹ [2026-02-11 03:58:13] Perform `Seurat::ScaleData()`
#> ℹ [2026-02-11 03:58:14] Perform pca linear dimension reduction
#> ℹ [2026-02-11 03:58:14] Perform `Seurat::FindClusters()` with `cluster_algorithm = 'louvain'` and `cluster_resolution = 0.6`
#> ℹ [2026-02-11 03:58:15] Reorder clusters...
#> ℹ [2026-02-11 03:58:15] Perform umap nonlinear dimension reduction
#> ℹ [2026-02-11 03:58:15] Non-linear dimensionality reduction (umap) using (Standardpca) dims (1-50) as input
#> ℹ [2026-02-11 03:58:19] Non-linear dimensionality reduction (umap) using (Standardpca) dims (1-50) as input
#> ✔ [2026-02-11 03:58:24] Run scop standard workflow completed
pancreas_sub <- RunSlingshot(
pancreas_sub,
group.by = "SubCellType",
reduction = "UMAP"
)
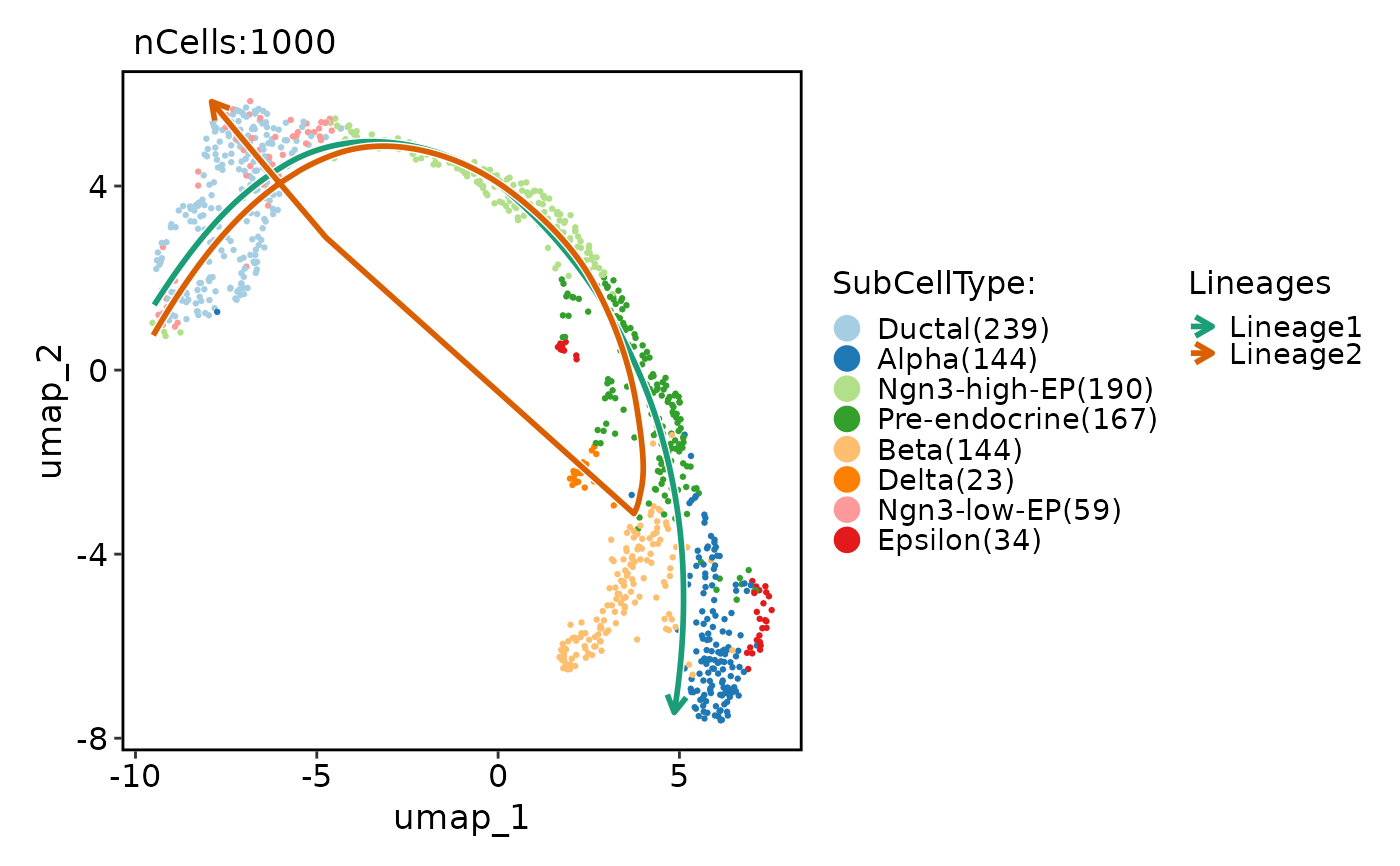 pancreas_sub <- RunDynamicFeatures(
pancreas_sub,
lineages = c("Lineage1", "Lineage2"),
n_candidates = 200
)
#> ℹ [2026-02-11 03:58:26] Start find dynamic features
#> ℹ [2026-02-11 03:58:26] Data type is raw counts
#> ℹ [2026-02-11 03:58:28] Number of candidate features (union): 231
#> ℹ [2026-02-11 03:58:28] Data type is raw counts
#> ℹ [2026-02-11 03:58:28] Calculating dynamic features for "Lineage1"...
#> ℹ [2026-02-11 03:58:28] Using 1 core
#> ⠙ [2026-02-11 03:58:28] Running for Gcg [1/231] ■ …
#> ⠹ [2026-02-11 03:58:28] Running for Gm8773 [87/231] ■■■■■■■■■■■■ …
#> ⠸ [2026-02-11 03:58:28] Running for BC048546 [173/231] ■■■■■■■■■■■■■■■■■■■■■■■ …
#> ✔ [2026-02-11 03:58:28] Completed 231 tasks in 8.1s
#>
#> ℹ [2026-02-11 03:58:28] Building results
#> ℹ [2026-02-11 03:58:37] Calculating dynamic features for "Lineage2"...
#> ℹ [2026-02-11 03:58:37] Using 1 core
#> ⠙ [2026-02-11 03:58:37] Running for Hmgb2 [25/231] ■■■■ …
#> ⠹ [2026-02-11 03:58:37] Running for Irx2 [107/231] ■■■■■■■■■■■■■■■ …
#> ⠸ [2026-02-11 03:58:37] Running for Tuba1a [184/231] ■■■■■■■■■■■■■■■■■■■■■■■■■ …
#> ✔ [2026-02-11 03:58:37] Completed 231 tasks in 8.6s
#>
#> ℹ [2026-02-11 03:58:37] Building results
#> ✔ [2026-02-11 03:58:45] Find dynamic features done
names(
pancreas_sub@tools$DynamicFeatures_Lineage1
)
#> [1] "DynamicFeatures" "raw_matrix" "fitted_matrix" "upr_matrix"
#> [5] "lwr_matrix" "libsize" "lineages" "family"
head(
pancreas_sub@tools$DynamicFeatures_Lineage1$DynamicFeatures
)
#> features exp_ncells r.sq dev.expl peaktime valleytime pvalue padjust
#> Gcg Gcg 182 0.6401140 0.7670504 21.33789 0.1164741 0 0
#> Ghrl Ghrl 167 0.2985260 0.6484728 18.01054 12.8984585 0 0
#> Iapp Iapp 279 0.2241776 0.7503415 21.33789 0.1164741 0 0
#> Pyy Pyy 434 0.4169149 0.7697879 19.21200 9.8743469 0 0
#> Rbp4 Rbp4 396 0.4403772 0.7285632 18.83423 10.1022997 0 0
#> Gast Gast 92 0.0659722 0.7077578 19.84490 0.1164741 0 0
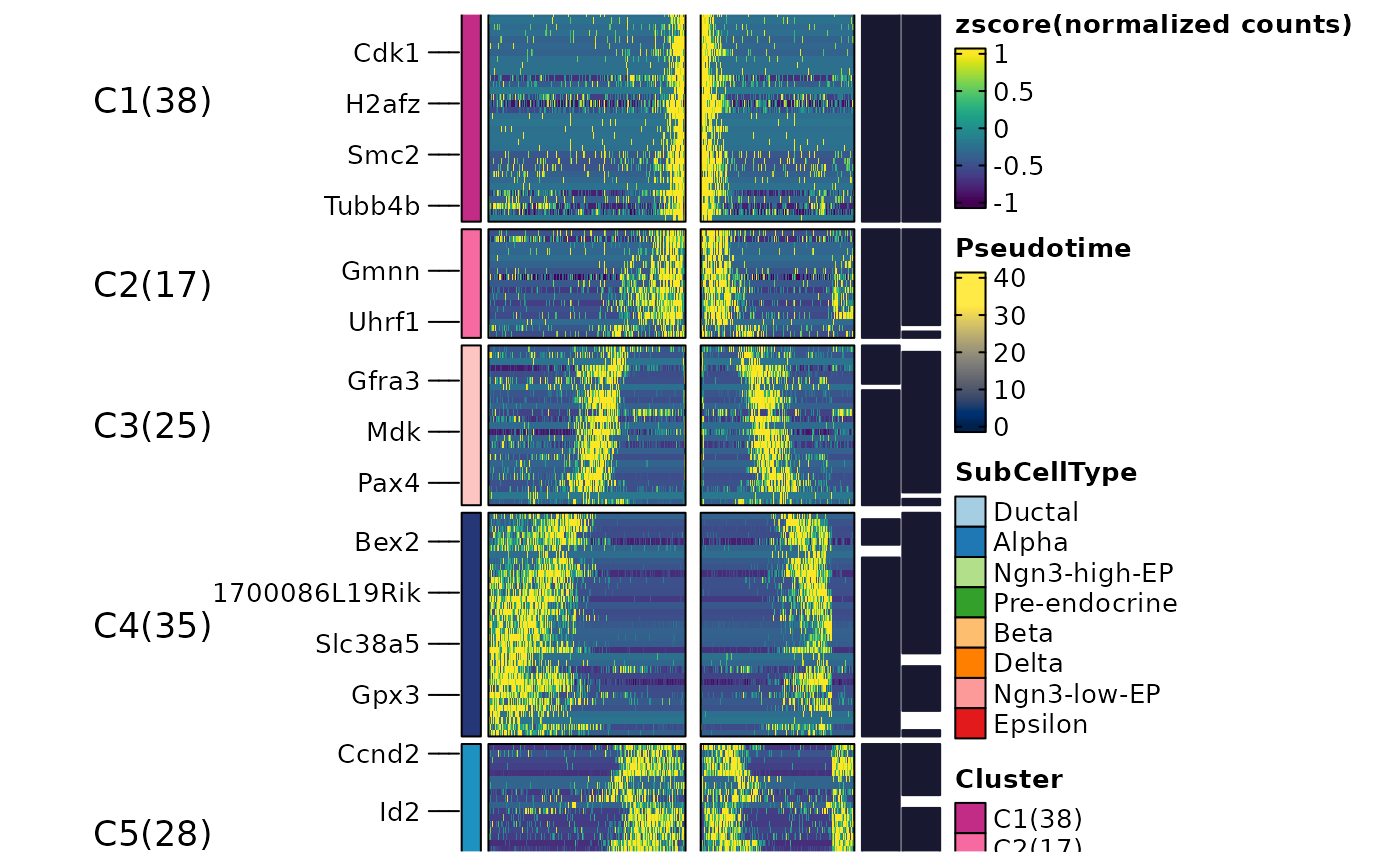
ht <- DynamicHeatmap(
pancreas_sub,
lineages = c("Lineage1", "Lineage2"),
cell_annotation = "SubCellType",
n_split = 6,
reverse_ht = "Lineage1"
)
#> ℹ [2026-02-11 03:58:45] [1] 180 features from Lineage1,Lineage2 passed the threshold (exp_ncells>[1] 20 & r.sq>[1] 0.2 & dev.expl>[1] 0.2 & padjust<[1] 0.05):
#> ℹ Gcg,Ghrl,Iapp,Pyy,Rbp4,Lrpprc,Slc38a5,Cdkn1a,2810417H13Rik,Chga...
#> ℹ [2026-02-11 03:58:47]
#> ℹ The size of the heatmap is fixed because certain elements are not scalable.
#> ℹ The width and height of the heatmap are determined by the size of the current viewport.
#> ℹ If you want to have more control over the size, you can manually set the parameters 'width' and 'height'.
pancreas_sub <- RunDynamicFeatures(
pancreas_sub,
lineages = c("Lineage1", "Lineage2"),
n_candidates = 200
)
#> ℹ [2026-02-11 03:58:26] Start find dynamic features
#> ℹ [2026-02-11 03:58:26] Data type is raw counts
#> ℹ [2026-02-11 03:58:28] Number of candidate features (union): 231
#> ℹ [2026-02-11 03:58:28] Data type is raw counts
#> ℹ [2026-02-11 03:58:28] Calculating dynamic features for "Lineage1"...
#> ℹ [2026-02-11 03:58:28] Using 1 core
#> ⠙ [2026-02-11 03:58:28] Running for Gcg [1/231] ■ …
#> ⠹ [2026-02-11 03:58:28] Running for Gm8773 [87/231] ■■■■■■■■■■■■ …
#> ⠸ [2026-02-11 03:58:28] Running for BC048546 [173/231] ■■■■■■■■■■■■■■■■■■■■■■■ …
#> ✔ [2026-02-11 03:58:28] Completed 231 tasks in 8.1s
#>
#> ℹ [2026-02-11 03:58:28] Building results
#> ℹ [2026-02-11 03:58:37] Calculating dynamic features for "Lineage2"...
#> ℹ [2026-02-11 03:58:37] Using 1 core
#> ⠙ [2026-02-11 03:58:37] Running for Hmgb2 [25/231] ■■■■ …
#> ⠹ [2026-02-11 03:58:37] Running for Irx2 [107/231] ■■■■■■■■■■■■■■■ …
#> ⠸ [2026-02-11 03:58:37] Running for Tuba1a [184/231] ■■■■■■■■■■■■■■■■■■■■■■■■■ …
#> ✔ [2026-02-11 03:58:37] Completed 231 tasks in 8.6s
#>
#> ℹ [2026-02-11 03:58:37] Building results
#> ✔ [2026-02-11 03:58:45] Find dynamic features done
names(
pancreas_sub@tools$DynamicFeatures_Lineage1
)
#> [1] "DynamicFeatures" "raw_matrix" "fitted_matrix" "upr_matrix"
#> [5] "lwr_matrix" "libsize" "lineages" "family"
head(
pancreas_sub@tools$DynamicFeatures_Lineage1$DynamicFeatures
)
#> features exp_ncells r.sq dev.expl peaktime valleytime pvalue padjust
#> Gcg Gcg 182 0.6401140 0.7670504 21.33789 0.1164741 0 0
#> Ghrl Ghrl 167 0.2985260 0.6484728 18.01054 12.8984585 0 0
#> Iapp Iapp 279 0.2241776 0.7503415 21.33789 0.1164741 0 0
#> Pyy Pyy 434 0.4169149 0.7697879 19.21200 9.8743469 0 0
#> Rbp4 Rbp4 396 0.4403772 0.7285632 18.83423 10.1022997 0 0
#> Gast Gast 92 0.0659722 0.7077578 19.84490 0.1164741 0 0
ht <- DynamicHeatmap(
pancreas_sub,
lineages = c("Lineage1", "Lineage2"),
cell_annotation = "SubCellType",
n_split = 6,
reverse_ht = "Lineage1"
)
#> ℹ [2026-02-11 03:58:45] [1] 180 features from Lineage1,Lineage2 passed the threshold (exp_ncells>[1] 20 & r.sq>[1] 0.2 & dev.expl>[1] 0.2 & padjust<[1] 0.05):
#> ℹ Gcg,Ghrl,Iapp,Pyy,Rbp4,Lrpprc,Slc38a5,Cdkn1a,2810417H13Rik,Chga...
#> ℹ [2026-02-11 03:58:47]
#> ℹ The size of the heatmap is fixed because certain elements are not scalable.
#> ℹ The width and height of the heatmap are determined by the size of the current viewport.
#> ℹ If you want to have more control over the size, you can manually set the parameters 'width' and 'height'.
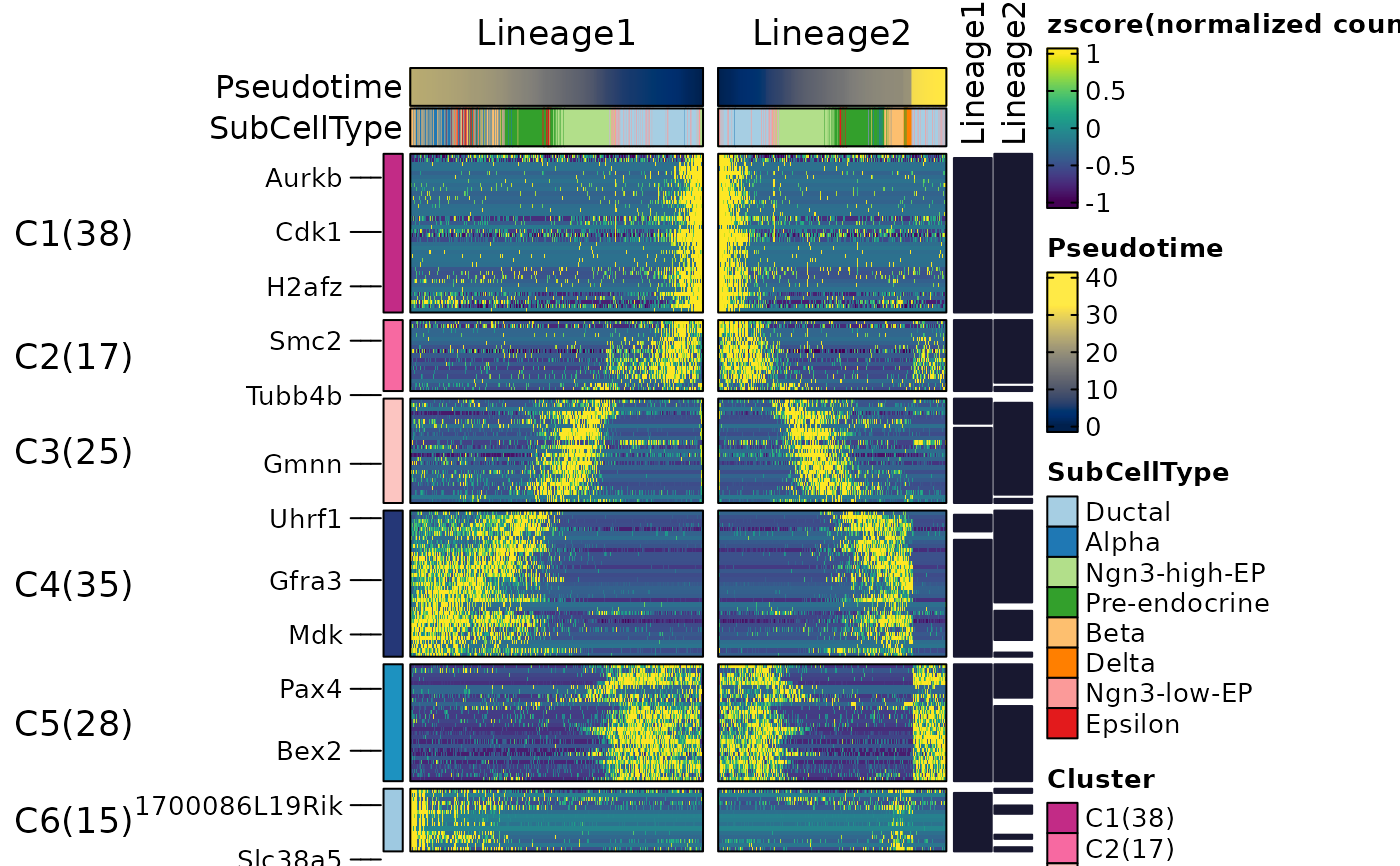 ht$plot
ht$plot
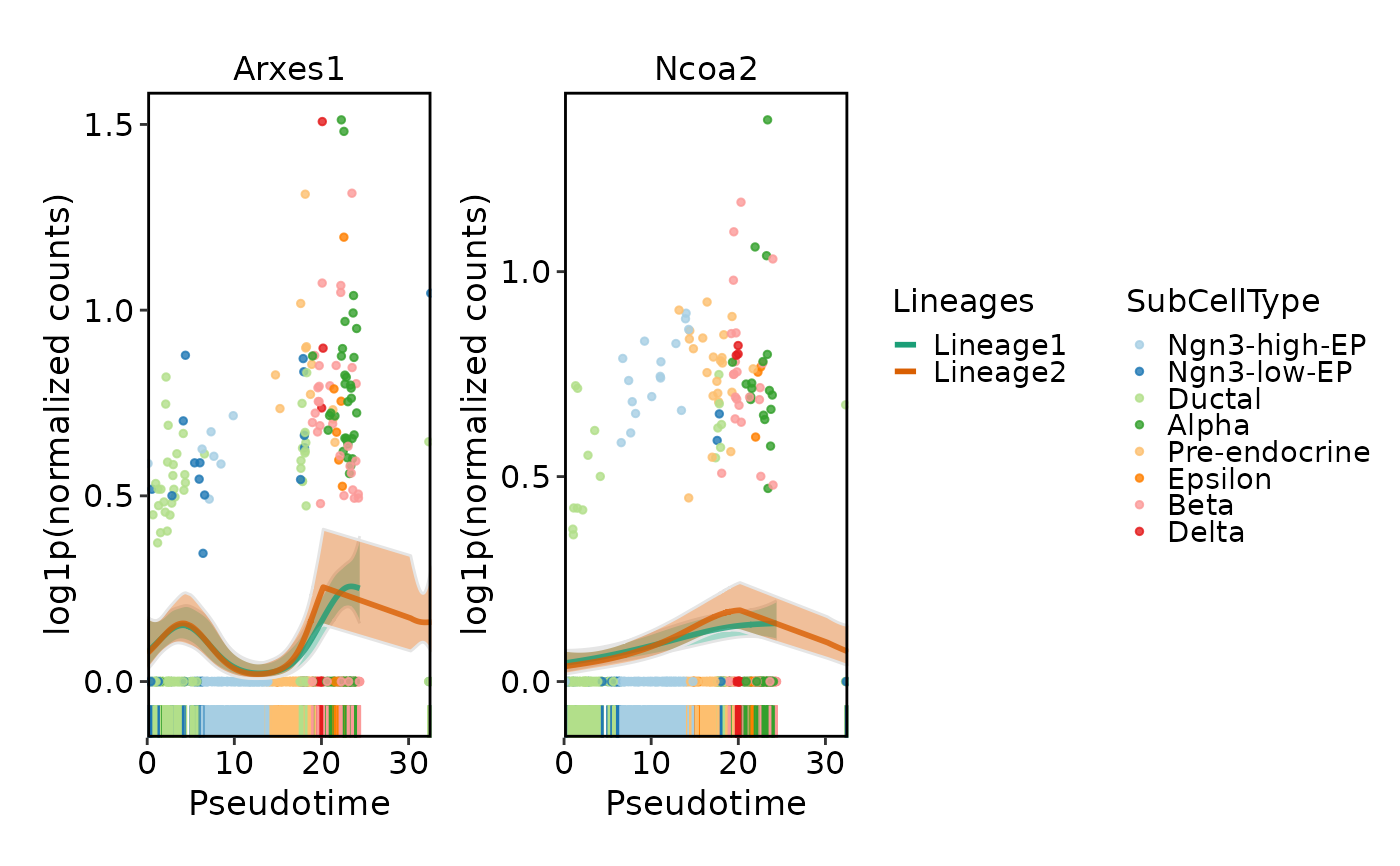
 DynamicPlot(
pancreas_sub,
lineages = c("Lineage1", "Lineage2"),
features = c("Arxes1", "Ncoa2"),
group.by = "SubCellType",
compare_lineages = TRUE,
compare_features = FALSE
)
#> ℹ [2026-02-11 03:58:50] Start find dynamic features
#> ℹ [2026-02-11 03:58:51] Data type is raw counts
#> ℹ [2026-02-11 03:58:51] Number of candidate features (union): 2
#> ℹ [2026-02-11 03:58:52] Data type is raw counts
#> ℹ [2026-02-11 03:58:52] Calculating dynamic features for "Lineage1"...
#> ℹ [2026-02-11 03:58:52] Using 1 core
#> ⠙ [2026-02-11 03:58:52] Running for Arxes1 [1/2] ■■■■■■■■■■■■■■■■ …
#> ✔ [2026-02-11 03:58:52] Completed 2 tasks in 152ms
#>
#> ℹ [2026-02-11 03:58:52] Building results
#> ✔ [2026-02-11 03:58:52] Find dynamic features done
#> ℹ [2026-02-11 03:58:52] Start find dynamic features
#> ℹ [2026-02-11 03:58:52] Data type is raw counts
#> ℹ [2026-02-11 03:58:53] Number of candidate features (union): 2
#> ℹ [2026-02-11 03:58:53] Data type is raw counts
#> ℹ [2026-02-11 03:58:53] Calculating dynamic features for "Lineage2"...
#> ℹ [2026-02-11 03:58:53] Using 1 core
#> ⠙ [2026-02-11 03:58:53] Running for Arxes1 [1/2] ■■■■■■■■■■■■■■■■ …
#> ✔ [2026-02-11 03:58:53] Completed 2 tasks in 127ms
#>
#> ℹ [2026-02-11 03:58:53] Building results
#> ✔ [2026-02-11 03:58:54] Find dynamic features done
DynamicPlot(
pancreas_sub,
lineages = c("Lineage1", "Lineage2"),
features = c("Arxes1", "Ncoa2"),
group.by = "SubCellType",
compare_lineages = TRUE,
compare_features = FALSE
)
#> ℹ [2026-02-11 03:58:50] Start find dynamic features
#> ℹ [2026-02-11 03:58:51] Data type is raw counts
#> ℹ [2026-02-11 03:58:51] Number of candidate features (union): 2
#> ℹ [2026-02-11 03:58:52] Data type is raw counts
#> ℹ [2026-02-11 03:58:52] Calculating dynamic features for "Lineage1"...
#> ℹ [2026-02-11 03:58:52] Using 1 core
#> ⠙ [2026-02-11 03:58:52] Running for Arxes1 [1/2] ■■■■■■■■■■■■■■■■ …
#> ✔ [2026-02-11 03:58:52] Completed 2 tasks in 152ms
#>
#> ℹ [2026-02-11 03:58:52] Building results
#> ✔ [2026-02-11 03:58:52] Find dynamic features done
#> ℹ [2026-02-11 03:58:52] Start find dynamic features
#> ℹ [2026-02-11 03:58:52] Data type is raw counts
#> ℹ [2026-02-11 03:58:53] Number of candidate features (union): 2
#> ℹ [2026-02-11 03:58:53] Data type is raw counts
#> ℹ [2026-02-11 03:58:53] Calculating dynamic features for "Lineage2"...
#> ℹ [2026-02-11 03:58:53] Using 1 core
#> ⠙ [2026-02-11 03:58:53] Running for Arxes1 [1/2] ■■■■■■■■■■■■■■■■ …
#> ✔ [2026-02-11 03:58:53] Completed 2 tasks in 127ms
#>
#> ℹ [2026-02-11 03:58:53] Building results
#> ✔ [2026-02-11 03:58:54] Find dynamic features done