This function calculates gene-set scores from the specified database (db) for each lineage using the specified scoring method (score_method).
It then treats these scores as expression values and uses them as input to the RunDynamicFeatures function to identify dynamically enriched terms along the lineage.
Usage
RunDynamicEnrichment(
srt,
lineages,
score_method = "AUCell",
layer = "data",
assay = NULL,
min_expcells = 20,
r.sq = 0.2,
dev.expl = 0.2,
padjust = 0.05,
IDtype = "symbol",
species = "Homo_sapiens",
db = "GO_BP",
db_update = FALSE,
db_version = "latest",
convert_species = TRUE,
Ensembl_version = NULL,
mirror = NULL,
TERM2GENE = NULL,
TERM2NAME = NULL,
minGSSize = 10,
maxGSSize = 500,
cores = 1,
verbose = TRUE,
seed = 11
)Arguments
- srt
A Seurat object containing the results of differential expression analysis (RunDEtest). If specified, the genes and groups will be extracted from the Seurat object automatically. If not specified, the
geneIDandgeneID_groupsarguments must be provided.- lineages
A character vector specifying the lineages to plot.
- score_method
The method to use for scoring. Can be
"Seurat","AUCell", or"UCell". Default is"Seurat".- layer
Which layer to use. Default is
"counts".- assay
Which assay to use. If
NULL, the default assay of the Seurat object will be used.- min_expcells
The minimum number of expected cells. Default is
20.- r.sq
The R-squared threshold. Default is
0.2.- dev.expl
The deviance explained threshold. Default is
0.2.- padjust
The p-value adjustment threshold. Default is
0.05.- IDtype
A character vector specifying the type of gene IDs in the
srtobject orgeneIDargument. This argument is used to convert the gene IDs to a different type ifIDtypeis different fromresult_IDtype.- species
A character vector specifying the species for which the gene annotation databases should be prepared. Can be
"Homo_sapiens"or"Mus_musculus".- db
A character vector specifying the annotation sources to be included in the gene annotation databases. Can be one or more of
"GO", "GO_BP", "GO_CC", "GO_MF", "KEGG", "WikiPathway", "Reactome", "CORUM", "MP", "DO", "HPO", "PFAM", "CSPA", "Surfaceome", "SPRomeDB", "VerSeDa", "TFLink", "hTFtarget", "TRRUST", "JASPAR", "ENCODE", "MSigDB", "CellTalk", "CellChat", "Chromosome", "GeneType", "Enzyme", "TF".- db_update
Whether the gene annotation databases should be forcefully updated. If set to FALSE, the function will attempt to load the cached databases instead. Default is
FALSE.- db_version
A character vector specifying the version of the gene annotation databases to be retrieved. Default is
"latest".- convert_species
Whether to use a species-converted database when the annotation is missing for the specified species. Default is
TRUE.- Ensembl_version
An integer specifying the Ensembl version. Default is
NULL. IfNULL, the latest version will be used.- mirror
Specify an Ensembl mirror to connect to. The valid options here are
"www","uswest","useast","asia".- TERM2GENE
A data frame specifying the gene-term mapping for a custom database. The first column should contain the term IDs, and the second column should contain the gene IDs.
- TERM2NAME
A data frame specifying the term-name mapping for a custom database. The first column should contain the term IDs, and the second column should contain the corresponding term names.
- minGSSize
The minimum size of a gene set to be considered in the enrichment analysis.
- maxGSSize
The maximum size of a gene set to be considered in the enrichment analysis.
- cores
The number of cores to use for parallelization with foreach::foreach. Default is
1.- verbose
Whether to print the message. Default is
TRUE.- seed
Random seed for reproducibility. Default is
11.
Examples
data(pancreas_sub)
pancreas_sub <- standard_scop(pancreas_sub)
#> ℹ [2026-02-11 03:53:56] Start standard scop workflow...
#> ℹ [2026-02-11 03:53:57] Checking a list of <Seurat>...
#> ! [2026-02-11 03:53:57] Data 1/1 of the `srt_list` is "unknown"
#> ℹ [2026-02-11 03:53:57] Perform `NormalizeData()` with `normalization.method = 'LogNormalize'` on the data 1/1 of the `srt_list`...
#> ℹ [2026-02-11 03:53:59] Perform `Seurat::FindVariableFeatures()` on the data 1/1 of the `srt_list`...
#> ℹ [2026-02-11 03:54:00] Use the separate HVF from srt_list
#> ℹ [2026-02-11 03:54:00] Number of available HVF: 2000
#> ℹ [2026-02-11 03:54:00] Finished check
#> ℹ [2026-02-11 03:54:00] Perform `Seurat::ScaleData()`
#> ℹ [2026-02-11 03:54:00] Perform pca linear dimension reduction
#> ℹ [2026-02-11 03:54:01] Perform `Seurat::FindClusters()` with `cluster_algorithm = 'louvain'` and `cluster_resolution = 0.6`
#> ℹ [2026-02-11 03:54:01] Reorder clusters...
#> ℹ [2026-02-11 03:54:02] Perform umap nonlinear dimension reduction
#> ℹ [2026-02-11 03:54:02] Non-linear dimensionality reduction (umap) using (Standardpca) dims (1-50) as input
#> ℹ [2026-02-11 03:54:06] Non-linear dimensionality reduction (umap) using (Standardpca) dims (1-50) as input
#> ✔ [2026-02-11 03:54:10] Run scop standard workflow completed
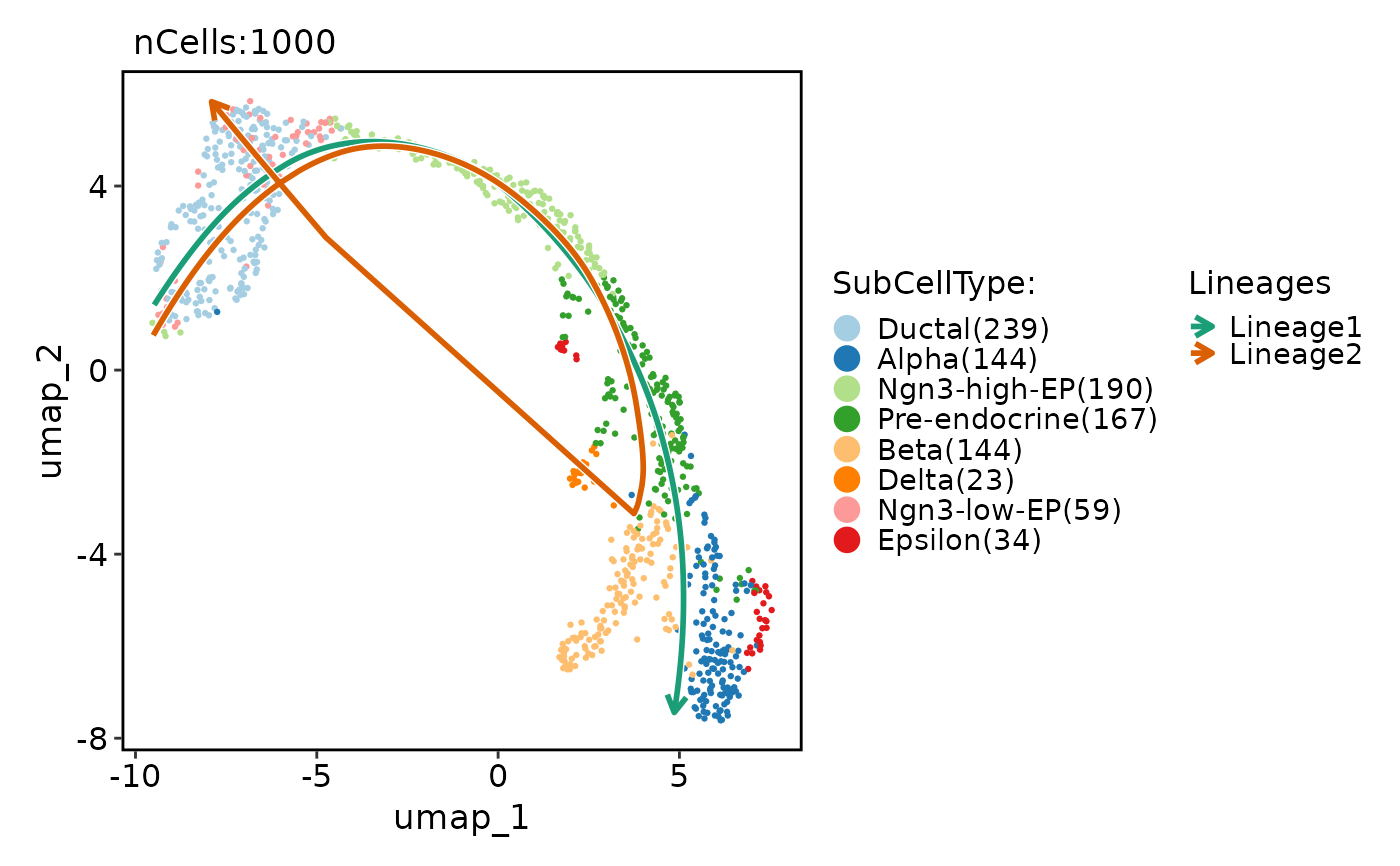
pancreas_sub <- RunSlingshot(
pancreas_sub,
group.by = "CellType",
reduction = "UMAP"
)
 pancreas_sub <- RunDynamicFeatures(
pancreas_sub,
lineages = "Lineage1",
n_candidates = 200
)
#> ℹ [2026-02-11 03:54:11] Start find dynamic features
#> ℹ [2026-02-11 03:54:12] Data type is raw counts
#> ℹ [2026-02-11 03:54:12] Number of candidate features (union): 200
#> ℹ [2026-02-11 03:54:13] Data type is raw counts
#> ℹ [2026-02-11 03:54:13] Calculating dynamic features for "Lineage1"...
#> ℹ [2026-02-11 03:54:13] Using 1 core
#> ⠙ [2026-02-11 03:54:13] Running for Gcg [1/200] ■ …
#> ⠹ [2026-02-11 03:54:13] Running for Lrpprc [10/200] ■■■ …
#> ⠸ [2026-02-11 03:54:13] Running for Avp [64/200] ■■■■■■■■■■■ …
#> ⠼ [2026-02-11 03:54:13] Running for Dbi [118/200] ■■■■■■■■■■■■■■■■■■■ …
#> ⠴ [2026-02-11 03:54:13] Running for Ascl1 [168/200] ■■■■■■■■■■■■■■■■■■■■■■■■■■ …
#> ✔ [2026-02-11 03:54:13] Completed 200 tasks in 11.5s
#>
#> ℹ [2026-02-11 03:54:13] Building results
#> ✔ [2026-02-11 03:54:24] Find dynamic features done
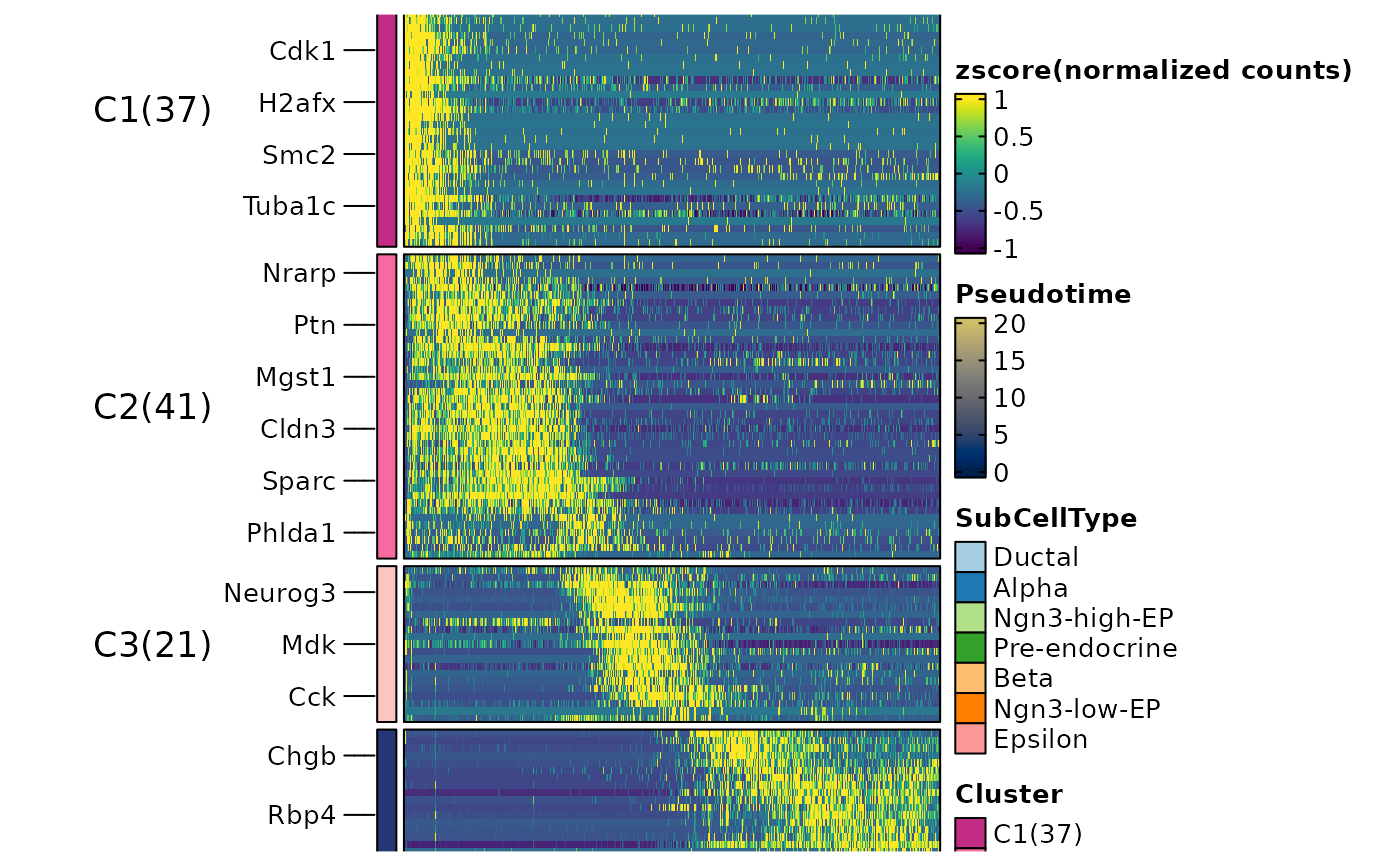
ht1 <- DynamicHeatmap(
pancreas_sub,
lineages = "Lineage1",
cell_annotation = "CellType",
n_split = 3
)
#> ℹ [2026-02-11 03:54:24] [1] 146 features from Lineage1 passed the threshold (exp_ncells>[1] 20 & r.sq>[1] 0.2 & dev.expl>[1] 0.2 & padjust<[1] 0.05):
#> ℹ Gcg,Ins1,Ins2,Nnat,Iapp,Lrpprc,Chgb,Slc38a5,2810417H13Rik,Rbp4...
#> ℹ [2026-02-11 03:54:25]
#> ℹ The size of the heatmap is fixed because certain elements are not scalable.
#> ℹ The width and height of the heatmap are determined by the size of the current viewport.
#> ℹ If you want to have more control over the size, you can manually set the parameters 'width' and 'height'.
pancreas_sub <- RunDynamicFeatures(
pancreas_sub,
lineages = "Lineage1",
n_candidates = 200
)
#> ℹ [2026-02-11 03:54:11] Start find dynamic features
#> ℹ [2026-02-11 03:54:12] Data type is raw counts
#> ℹ [2026-02-11 03:54:12] Number of candidate features (union): 200
#> ℹ [2026-02-11 03:54:13] Data type is raw counts
#> ℹ [2026-02-11 03:54:13] Calculating dynamic features for "Lineage1"...
#> ℹ [2026-02-11 03:54:13] Using 1 core
#> ⠙ [2026-02-11 03:54:13] Running for Gcg [1/200] ■ …
#> ⠹ [2026-02-11 03:54:13] Running for Lrpprc [10/200] ■■■ …
#> ⠸ [2026-02-11 03:54:13] Running for Avp [64/200] ■■■■■■■■■■■ …
#> ⠼ [2026-02-11 03:54:13] Running for Dbi [118/200] ■■■■■■■■■■■■■■■■■■■ …
#> ⠴ [2026-02-11 03:54:13] Running for Ascl1 [168/200] ■■■■■■■■■■■■■■■■■■■■■■■■■■ …
#> ✔ [2026-02-11 03:54:13] Completed 200 tasks in 11.5s
#>
#> ℹ [2026-02-11 03:54:13] Building results
#> ✔ [2026-02-11 03:54:24] Find dynamic features done
ht1 <- DynamicHeatmap(
pancreas_sub,
lineages = "Lineage1",
cell_annotation = "CellType",
n_split = 3
)
#> ℹ [2026-02-11 03:54:24] [1] 146 features from Lineage1 passed the threshold (exp_ncells>[1] 20 & r.sq>[1] 0.2 & dev.expl>[1] 0.2 & padjust<[1] 0.05):
#> ℹ Gcg,Ins1,Ins2,Nnat,Iapp,Lrpprc,Chgb,Slc38a5,2810417H13Rik,Rbp4...
#> ℹ [2026-02-11 03:54:25]
#> ℹ The size of the heatmap is fixed because certain elements are not scalable.
#> ℹ The width and height of the heatmap are determined by the size of the current viewport.
#> ℹ If you want to have more control over the size, you can manually set the parameters 'width' and 'height'.
 pancreas_sub <- RunDynamicEnrichment(
pancreas_sub,
lineages = "Lineage1",
score_method = "AUCell",
db = "GO_BP",
species = "Mus_musculus"
)
#> ℹ [2026-02-11 03:54:27] Species: "Mus_musculus"
#> ℹ [2026-02-11 03:54:27] Loading cached: GO_BP version: 3.22.0 nterm:15169 created: 2026-02-11 03:27:59
#> ℹ [2026-02-11 03:54:29] Start cell scoring
#> ℹ [2026-02-11 03:54:30] Data type is log-normalized
#> ℹ [2026-02-11 03:54:31] Number of feature lists to be scored: 2761
#> ✔ [2026-02-11 03:57:18] Cell scoring completed
#> ℹ [2026-02-11 03:57:18] Start find dynamic features
#> ℹ [2026-02-11 03:57:18] Data type is log-normalized
#> ℹ [2026-02-11 03:57:18] Number of candidate features (union): 2761
#> ℹ [2026-02-11 03:57:18] Data type is log-normalized
#> ℹ [2026-02-11 03:57:19] Calculating dynamic features for "Lineage1"...
#> ℹ [2026-02-11 03:57:19] Using 1 core
#> ⠙ [2026-02-11 03:57:19] Running for GO-BP-2..deoxyribonucleotide.biosynthetic.p…
#> ⠹ [2026-02-11 03:57:19] Running for GO-BP-RNA.localization [56/2761] ■■ …
#> ⠸ [2026-02-11 03:57:19] Running for GO-BP-cartilage.condensation [268/2761] ■■■…
#> ⠼ [2026-02-11 03:57:19] Running for GO-BP-detection.of.light.stimulus.involved.…
#> ⠴ [2026-02-11 03:57:19] Running for GO-BP-heart.contraction [690/2761] ■■■■■■■■…
#> ⠦ [2026-02-11 03:57:19] Running for GO-BP-megakaryocyte.differentiation [901/27…
#> ⠧ [2026-02-11 03:57:19] Running for GO-BP-negative.regulation.of.cell.developme…
#> ⠇ [2026-02-11 03:57:19] Running for GO-BP-neutral.amino.acid.transport [1332/27…
#> ⠏ [2026-02-11 03:57:19] Running for GO-BP-positive.regulation.of.cell.substrate…
#> ⠋ [2026-02-11 03:57:19] Running for GO-BP-positive.regulation.of.telomere.maint…
#> ⠙ [2026-02-11 03:57:19] Running for GO-BP-regulation.of.animal.organ.morphogene…
#> ⠹ [2026-02-11 03:57:19] Running for GO-BP-regulation.of.lymphocyte.apoptotic.pr…
#> ⠸ [2026-02-11 03:57:19] Running for GO-BP-regulation.of.system.process [2383/27…
#> ⠼ [2026-02-11 03:57:19] Running for GO-BP-skin.epidermis.development [2596/2761…
#> ✔ [2026-02-11 03:57:19] Completed 2761 tasks in 39.1s
#>
#> ℹ [2026-02-11 03:57:19] Building results
#> ✔ [2026-02-11 03:57:58] Find dynamic features done
#> ✔ [2026-02-11 03:57:58] Dynamic enrichment analysis completed
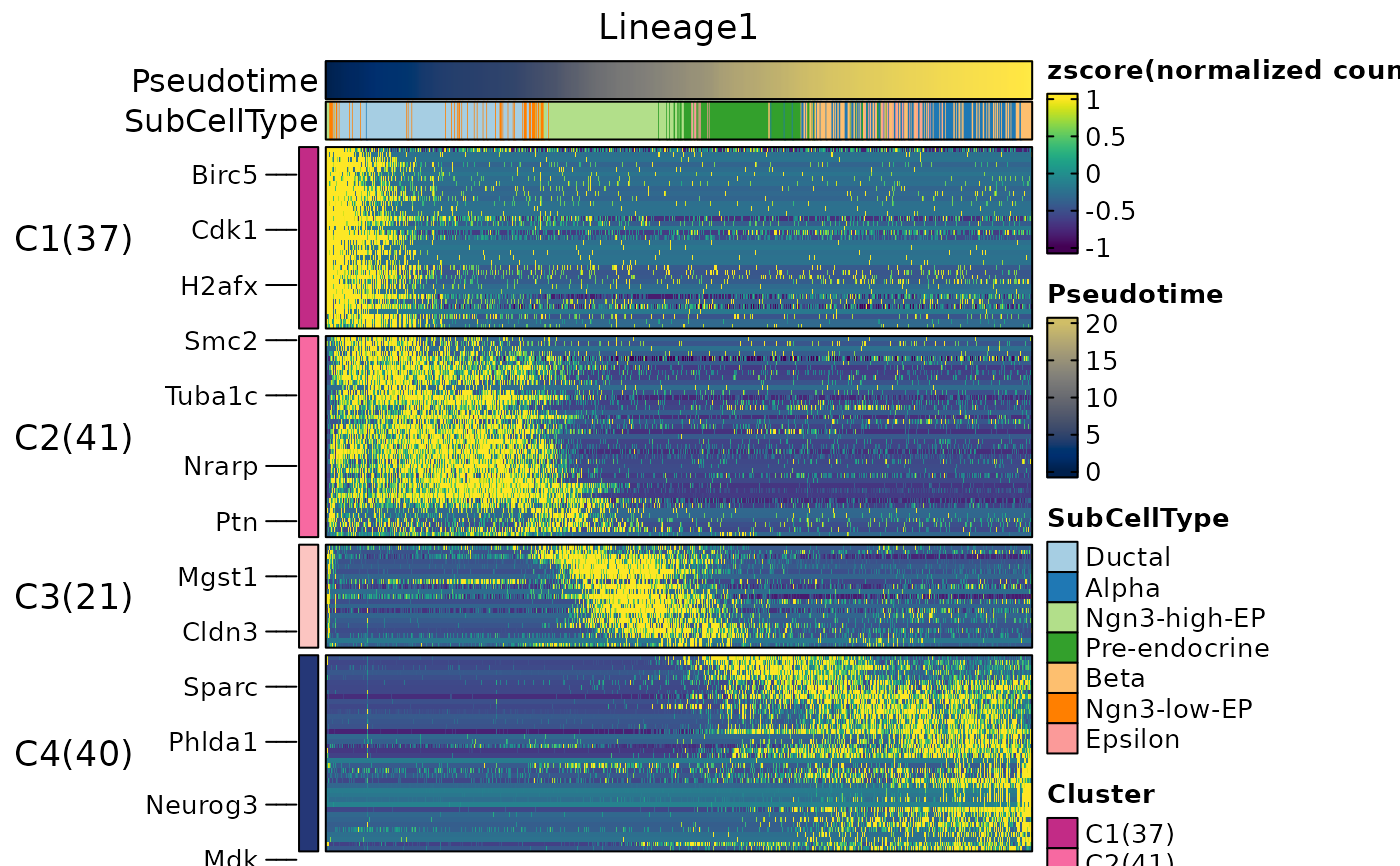
ht2 <- DynamicHeatmap(
pancreas_sub,
assay = "GO_BP",
lineages = "Lineage1_GO_BP",
cell_annotation = "CellType",
n_split = 3,
split_method = "kmeans-peaktime"
)
#> ℹ [2026-02-11 03:57:58] [1] 1897 features from Lineage1_GO_BP passed the threshold (exp_ncells>[1] 20 & r.sq>[1] 0.2 & dev.expl>[1] 0.2 & padjust<[1] 0.05):
#> ℹ GO-BP-2..deoxyribonucleotide.biosynthetic.process,GO-BP-2..deoxyribonucleotide.metabolic.process,GO-BP-ADP.catabolic.process,GO-BP-ADP.metabolic.process,GO-BP-ATP.biosynthetic.process,GO-BP-ATP.metabolic.process,GO-BP-ATP.synthesis.coupled.electron.transport,GO-BP-B.cell.activation,GO-BP-B.cell.apoptotic.process,GO-BP-B.cell.proliferation...
#> ! [2026-02-11 03:57:58] The values in the 'counts' layer are non-integer. Set the library size to 1.
#> ℹ [2026-02-11 03:57:59]
#> ℹ The size of the heatmap is fixed because certain elements are not scalable.
#> ℹ The width and height of the heatmap are determined by the size of the current viewport.
#> ℹ If you want to have more control over the size, you can manually set the parameters 'width' and 'height'.
pancreas_sub <- RunDynamicEnrichment(
pancreas_sub,
lineages = "Lineage1",
score_method = "AUCell",
db = "GO_BP",
species = "Mus_musculus"
)
#> ℹ [2026-02-11 03:54:27] Species: "Mus_musculus"
#> ℹ [2026-02-11 03:54:27] Loading cached: GO_BP version: 3.22.0 nterm:15169 created: 2026-02-11 03:27:59
#> ℹ [2026-02-11 03:54:29] Start cell scoring
#> ℹ [2026-02-11 03:54:30] Data type is log-normalized
#> ℹ [2026-02-11 03:54:31] Number of feature lists to be scored: 2761
#> ✔ [2026-02-11 03:57:18] Cell scoring completed
#> ℹ [2026-02-11 03:57:18] Start find dynamic features
#> ℹ [2026-02-11 03:57:18] Data type is log-normalized
#> ℹ [2026-02-11 03:57:18] Number of candidate features (union): 2761
#> ℹ [2026-02-11 03:57:18] Data type is log-normalized
#> ℹ [2026-02-11 03:57:19] Calculating dynamic features for "Lineage1"...
#> ℹ [2026-02-11 03:57:19] Using 1 core
#> ⠙ [2026-02-11 03:57:19] Running for GO-BP-2..deoxyribonucleotide.biosynthetic.p…
#> ⠹ [2026-02-11 03:57:19] Running for GO-BP-RNA.localization [56/2761] ■■ …
#> ⠸ [2026-02-11 03:57:19] Running for GO-BP-cartilage.condensation [268/2761] ■■■…
#> ⠼ [2026-02-11 03:57:19] Running for GO-BP-detection.of.light.stimulus.involved.…
#> ⠴ [2026-02-11 03:57:19] Running for GO-BP-heart.contraction [690/2761] ■■■■■■■■…
#> ⠦ [2026-02-11 03:57:19] Running for GO-BP-megakaryocyte.differentiation [901/27…
#> ⠧ [2026-02-11 03:57:19] Running for GO-BP-negative.regulation.of.cell.developme…
#> ⠇ [2026-02-11 03:57:19] Running for GO-BP-neutral.amino.acid.transport [1332/27…
#> ⠏ [2026-02-11 03:57:19] Running for GO-BP-positive.regulation.of.cell.substrate…
#> ⠋ [2026-02-11 03:57:19] Running for GO-BP-positive.regulation.of.telomere.maint…
#> ⠙ [2026-02-11 03:57:19] Running for GO-BP-regulation.of.animal.organ.morphogene…
#> ⠹ [2026-02-11 03:57:19] Running for GO-BP-regulation.of.lymphocyte.apoptotic.pr…
#> ⠸ [2026-02-11 03:57:19] Running for GO-BP-regulation.of.system.process [2383/27…
#> ⠼ [2026-02-11 03:57:19] Running for GO-BP-skin.epidermis.development [2596/2761…
#> ✔ [2026-02-11 03:57:19] Completed 2761 tasks in 39.1s
#>
#> ℹ [2026-02-11 03:57:19] Building results
#> ✔ [2026-02-11 03:57:58] Find dynamic features done
#> ✔ [2026-02-11 03:57:58] Dynamic enrichment analysis completed
ht2 <- DynamicHeatmap(
pancreas_sub,
assay = "GO_BP",
lineages = "Lineage1_GO_BP",
cell_annotation = "CellType",
n_split = 3,
split_method = "kmeans-peaktime"
)
#> ℹ [2026-02-11 03:57:58] [1] 1897 features from Lineage1_GO_BP passed the threshold (exp_ncells>[1] 20 & r.sq>[1] 0.2 & dev.expl>[1] 0.2 & padjust<[1] 0.05):
#> ℹ GO-BP-2..deoxyribonucleotide.biosynthetic.process,GO-BP-2..deoxyribonucleotide.metabolic.process,GO-BP-ADP.catabolic.process,GO-BP-ADP.metabolic.process,GO-BP-ATP.biosynthetic.process,GO-BP-ATP.metabolic.process,GO-BP-ATP.synthesis.coupled.electron.transport,GO-BP-B.cell.activation,GO-BP-B.cell.apoptotic.process,GO-BP-B.cell.proliferation...
#> ! [2026-02-11 03:57:58] The values in the 'counts' layer are non-integer. Set the library size to 1.
#> ℹ [2026-02-11 03:57:59]
#> ℹ The size of the heatmap is fixed because certain elements are not scalable.
#> ℹ The width and height of the heatmap are determined by the size of the current viewport.
#> ℹ If you want to have more control over the size, you can manually set the parameters 'width' and 'height'.