Heatmap plot for dynamic features along lineages
Usage
DynamicHeatmap(
srt,
lineages,
features = NULL,
use_fitted = FALSE,
border = TRUE,
flip = FALSE,
min_expcells = 20,
r.sq = 0.2,
dev.expl = 0.2,
padjust = 0.05,
num_intersections = NULL,
cell_density = 1,
cell_bins = 100,
order_by = c("peaktime", "valleytime"),
layer = "counts",
assay = NULL,
exp_method = c("zscore", "raw", "fc", "log2fc", "log1p"),
exp_legend_title = NULL,
limits = NULL,
lib_normalize = identical(layer, "counts"),
libsize = NULL,
family = NULL,
cluster_features_by = NULL,
cluster_rows = FALSE,
cluster_row_slices = FALSE,
cluster_columns = FALSE,
cluster_column_slices = FALSE,
show_row_names = FALSE,
show_column_names = FALSE,
row_names_side = ifelse(flip, "left", "right"),
column_names_side = ifelse(flip, "bottom", "top"),
row_names_rot = 0,
column_names_rot = 90,
row_title = NULL,
column_title = NULL,
row_title_side = "left",
column_title_side = "top",
row_title_rot = 0,
column_title_rot = ifelse(flip, 90, 0),
feature_split = NULL,
feature_split_by = NULL,
n_split = NULL,
split_order = NULL,
split_method = c("mfuzz", "kmeans", "kmeans-peaktime", "hclust", "hclust-peaktime"),
decreasing = FALSE,
fuzzification = NULL,
anno_terms = FALSE,
anno_keys = FALSE,
anno_features = FALSE,
terms_width = grid::unit(4, "in"),
terms_fontsize = 8,
keys_width = grid::unit(2, "in"),
keys_fontsize = c(6, 10),
features_width = grid::unit(2, "in"),
features_fontsize = c(6, 10),
IDtype = "symbol",
species = "Homo_sapiens",
db_update = FALSE,
db_version = "latest",
db_combine = FALSE,
convert_species = FALSE,
Ensembl_version = NULL,
mirror = NULL,
db = "GO_BP",
TERM2GENE = NULL,
TERM2NAME = NULL,
minGSSize = 10,
maxGSSize = 500,
GO_simplify = FALSE,
GO_simplify_cutoff = "p.adjust < 0.05",
simplify_method = "Wang",
simplify_similarityCutoff = 0.7,
pvalueCutoff = NULL,
padjustCutoff = 0.05,
topTerm = 5,
show_termid = FALSE,
topWord = 20,
words_excluded = NULL,
nlabel = 20,
features_label = NULL,
label_size = 10,
label_color = "black",
pseudotime_label = NULL,
pseudotime_label_color = "black",
pseudotime_label_linetype = 2,
pseudotime_label_linewidth = 3,
heatmap_palette = "viridis",
heatmap_palcolor = NULL,
pseudotime_palette = "cividis",
pseudotime_palcolor = NULL,
feature_split_palette = "simspec",
feature_split_palcolor = NULL,
cell_annotation = NULL,
cell_annotation_palette = "Paired",
cell_annotation_palcolor = NULL,
cell_annotation_params = if (flip) {
list(width = grid::unit(5, "mm"))
} else {
list(height = grid::unit(5, "mm"))
},
feature_annotation = NULL,
feature_annotation_palette = "Dark2",
feature_annotation_palcolor = NULL,
feature_annotation_params = if (flip) {
list(height = grid::unit(5, "mm"))
} else
{
list(width = grid::unit(5, "mm"))
},
separate_annotation = NULL,
separate_annotation_palette = "Paired",
separate_annotation_palcolor = NULL,
separate_annotation_params = if (flip) {
list(width = grid::unit(10, "mm"))
}
else {
list(height = grid::unit(10, "mm"))
},
reverse_ht = NULL,
use_raster = NULL,
raster_device = "png",
raster_by_magick = TRUE,
height = NULL,
width = NULL,
units = "inch",
seed = 11,
ht_params = list()
)Arguments
- srt
A Seurat object.
- lineages
A character vector specifying the lineages to plot.
- features
A character vector specifying the features to plot. By default, this parameter is set to NULL, and the dynamic features will be determined by the parameters
min_expcells,r.sq,dev.expl,padjustandnum_intersections.- use_fitted
Whether to use fitted values. Default is
FALSE.- border
Whether to add a border to the heatmap. Default is
TRUE.- flip
Whether to flip the heatmap. Default is
FALSE.- min_expcells
The minimum number of expected cells. Default is
20.- r.sq
The R-squared threshold. Default is
0.2.- dev.expl
The deviance explained threshold. Default is
0.2.- padjust
The p-value adjustment threshold. Default is
0.05.- num_intersections
This parameter is a numeric vector used to determine the number of intersections among lineages. It helps in selecting which dynamic features will be used. By default, when this parameter is set to
NULL, all dynamic features that pass the specified threshold will be used for each lineage.- cell_density
The cell density within each cell bin. By default, this parameter is set to
1, which means that all cells will be included within each cell bin.- cell_bins
The number of cell bins. Default is
100.- order_by
The order of the heatmap. Default is
"peaktime".- layer
Which layer to use. Default is
"counts".- assay
Which assay to use. If
NULL, the default assay of the Seurat object will be used.- exp_method
A character vector specifying the method for calculating expression values. Options are
"zscore","raw","fc","log2fc", or"log1p". Default is"zscore".- exp_legend_title
A character vector specifying the title for the legend of expression value. Default is
NULL.- limits
A two-length numeric vector specifying the limits for the color scale. Default is
NULL.- lib_normalize
Whether to normalize the data by library size.
- libsize
A numeric vector specifying the library size for each cell. Default is
NULL.- family
The model used to calculate the dynamic features if needed. By default, this parameter is set to
NULL, and the appropriate family will be automatically determined.- cluster_features_by
Which lineage to use when clustering features. By default, this parameter is set to
NULL, which means that all lineages will be used.- cluster_rows
Whether to cluster rows in the heatmap. Default is
FALSE.- cluster_row_slices
Whether to cluster row slices in the heatmap. Default is
FALSE.- cluster_columns
Whether to cluster columns in the heatmap. Default is
FALSE.- cluster_column_slices
Whether to cluster column slices in the heatmap. Default is
FALSE.- show_row_names
Whether to show row names in the heatmap. Default is
FALSE.- show_column_names
Whether to show column names in the heatmap. Default is
FALSE.- row_names_side
A character vector specifying the side to place row names.
- column_names_side
A character vector specifying the side to place column names.
- row_names_rot
The rotation angle for row names. Default is
0.- column_names_rot
The rotation angle for column names. Default is
90.- row_title
A character vector specifying the title for rows. Default is
NULL.- column_title
A character vector specifying the title for columns. Default is
NULL.- row_title_side
A character vector specifying the side to place row title. Default is
"left".- column_title_side
A character vector specifying the side to place column title. Default is
"top".- row_title_rot
The rotation angle for row title. Default is
0.- column_title_rot
The rotation angle for column title.
- feature_split
A factor specifying how to split the features. Default is
NULL.- feature_split_by
A character vector specifying which group.by to use when splitting features (into n_split feature clusters). Default is
NULL.- n_split
A number of feature splits (feature clusters) to create. Default is
NULL.- split_order
A numeric vector specifying the order of splits. Default is
NULL.- split_method
A character vector specifying the method for splitting features. Options are
"kmeans","hclust", or"mfuzz". Default is"kmeans".- decreasing
Whether to sort feature splits in decreasing order. Default is
FALSE.- fuzzification
The fuzzification coefficient. Default is
NULL.- anno_terms
Whether to include term annotations. Default is
FALSE.- anno_keys
Whether to include key annotations. Default is
FALSE.- anno_features
Whether to include feature annotations. Default is
FALSE.- terms_width
A unit specifying the width of term annotations. Default is
unit(4, "in").- terms_fontsize
A numeric vector specifying the font size(s) for term annotations. Default is
8.- keys_width
A unit specifying the width of key annotations. Default is
unit(2, "in").- keys_fontsize
A two-length numeric vector specifying the minimum and maximum font size(s) for key annotations. Default is
c(6, 10).- features_width
A unit specifying the width of feature annotations. Default is
unit(2, "in").- features_fontsize
A two-length numeric vector specifying the minimum and maximum font size(s) for feature annotations. Default is
c(6, 10).- IDtype
A character vector specifying the type of IDs for features. Default is
"symbol".- species
A character vector specifying the species for features. Default is
"Homo_sapiens".- db_update
Whether the gene annotation databases should be forcefully updated. If set to FALSE, the function will attempt to load the cached databases instead. Default is
FALSE.- db_version
A character vector specifying the version of the gene annotation databases to be retrieved. Default is
"latest".- db_combine
Whether to use a combined database. Default is
FALSE.- convert_species
Whether to use a species-converted database when the annotation is missing for the specified species. Default is
TRUE.- Ensembl_version
An integer specifying the Ensembl version. Default is
NULL. IfNULL, the latest version will be used.- mirror
A character vector specifying the mirror for the Ensembl database. Default is
NULL.- db
A character vector specifying the database to use. Default is
"GO_BP".- TERM2GENE
A data.frame specifying the TERM2GENE mapping for the database. Default is
NULL.- TERM2NAME
A data.frame specifying the TERM2NAME mapping for the database. Default is
NULL.- minGSSize
An integer specifying the minimum gene set size for the database. Default is
10.- maxGSSize
An integer specifying the maximum gene set size for the database. Default is
500.- GO_simplify
Whether to simplify gene ontology terms. Default is
FALSE.- GO_simplify_cutoff
A character vector specifying the cutoff for GO simplification. Default is
"p.adjust < 0.05".- simplify_method
A character vector specifying the method for GO simplification. Default is
"Wang".- simplify_similarityCutoff
The similarity cutoff for GO simplification. Default is
0.7.- pvalueCutoff
A numeric vector specifying the p-value cutoff(s) for significance. Default is
NULL.- padjustCutoff
The adjusted p-value cutoff for significance. Default is
0.05.- topTerm
A number of top terms to include. Default is
5.- show_termid
Whether to show term IDs. Default is
FALSE.- topWord
A number of top words to include. Default is
20.- words_excluded
A character vector specifying the words to exclude. Default is
NULL.- nlabel
A number of labels to include. Default is
20.- features_label
A character vector specifying the features to label. Default is
NULL.- label_size
The size of labels. Default is
10.- label_color
A character vector specifying the color of labels. Default is
"black".- pseudotime_label
The pseudotime label. Default is
NULL.- pseudotime_label_color
The pseudotime label color. Default is
"black".- pseudotime_label_linetype
The pseudotime label line type. Default is
2.- pseudotime_label_linewidth
The pseudotime label line width. Default is
3.- heatmap_palette
A character vector specifying the palette to use for the heatmap. Default is
"RdBu".- heatmap_palcolor
A character vector specifying the heatmap color to use. Default is
NULL.- pseudotime_palette
The color palette to use for pseudotime. Default is
"cividis".- pseudotime_palcolor
The colors to use for the pseudotime in the heatmap. Default is
NULL.- feature_split_palette
A character vector specifying the palette to use for feature splits. Default is
"simspec".- feature_split_palcolor
A character vector specifying the feature split color to use. Default is
NULL.- cell_annotation
A character vector specifying the cell annotation(s) to include. Default is
NULL.- cell_annotation_palette
A character vector specifying the palette to use for cell annotations. The length of the vector should match the number of cell_annotation. Default is
"Paired".- cell_annotation_palcolor
A list of character vector specifying the cell annotation color(s) to use. The length of the list should match the number of cell_annotation. Default is
NULL.- cell_annotation_params
A list specifying additional parameters for cell annotations. Default is a list with
width = unit(1, "cm")if flip is TRUE, else a list withheight = unit(1, "cm").- feature_annotation
A character vector specifying the feature annotation(s) to include. Default is
NULL.- feature_annotation_palette
A character vector specifying the palette to use for feature annotations. The length of the vector should match the number of feature_annotation. Default is
"Dark2".- feature_annotation_palcolor
A list of character vector specifying the feature annotation color to use. The length of the list should match the number of feature_annotation. Default is
NULL.- feature_annotation_params
A list specifying additional parameters for feature annotations. Default is
list().- separate_annotation
Names of the annotations to be displayed in separate annotation blocks. Each name should match a column name in the metadata of the
Seuratobject.- separate_annotation_palette
The color palette to use for separate annotations. Default is
"Paired".- separate_annotation_palcolor
The colors to use for each level of the separate annotations. Default is
NULL.- separate_annotation_params
Other parameters to ComplexHeatmap::HeatmapAnnotation when creating a separate annotation blocks.
- reverse_ht
Whether to reverse the heatmap. Default is
NULL.- use_raster
Whether to use a raster device for plotting. Default is
NULL.- raster_device
A character vector specifying the raster device to use. Default is
"png".- raster_by_magick
Whether to use the 'magick' package for raster. Default is
FALSE.- height
A numeric vector specifying the height(s) of the heatmap body. Default is
NULL.- width
A numeric vector specifying the width(s) of the heatmap body. Default is
NULL.- units
A character vector specifying the units for the height and width. Default is
"inch".- seed
Random seed for reproducibility. Default is
11.- ht_params
Additional parameters to customize the appearance of the heatmap. This should be a list with named elements, where the names correspond to parameter names in the ComplexHeatmap::Heatmap function. Any conflicting parameters will override the defaults set by this function. Default is
list().
Examples
data(pancreas_sub)
pancreas_sub <- standard_scop(pancreas_sub)
#> ℹ [2026-02-11 03:16:59] Start standard scop workflow...
#> ℹ [2026-02-11 03:17:00] Checking a list of <Seurat>...
#> ! [2026-02-11 03:17:00] Data 1/1 of the `srt_list` is "unknown"
#> ℹ [2026-02-11 03:17:00] Perform `NormalizeData()` with `normalization.method = 'LogNormalize'` on the data 1/1 of the `srt_list`...
#> ℹ [2026-02-11 03:17:01] Perform `Seurat::FindVariableFeatures()` on the data 1/1 of the `srt_list`...
#> ℹ [2026-02-11 03:17:02] Use the separate HVF from srt_list
#> ℹ [2026-02-11 03:17:02] Number of available HVF: 2000
#> ℹ [2026-02-11 03:17:02] Finished check
#> ℹ [2026-02-11 03:17:02] Perform `Seurat::ScaleData()`
#> ℹ [2026-02-11 03:17:03] Perform pca linear dimension reduction
#> ℹ [2026-02-11 03:17:04] Perform `Seurat::FindClusters()` with `cluster_algorithm = 'louvain'` and `cluster_resolution = 0.6`
#> ℹ [2026-02-11 03:17:04] Reorder clusters...
#> ℹ [2026-02-11 03:17:04] Perform umap nonlinear dimension reduction
#> ℹ [2026-02-11 03:17:04] Non-linear dimensionality reduction (umap) using (Standardpca) dims (1-50) as input
#> ℹ [2026-02-11 03:17:07] Non-linear dimensionality reduction (umap) using (Standardpca) dims (1-50) as input
#> ✔ [2026-02-11 03:17:10] Run scop standard workflow completed
pancreas_sub <- RunSlingshot(
pancreas_sub,
group.by = "SubCellType",
reduction = "UMAP"
)
#> Warning: No shared levels found between `names(values)` of the manual scale and the
#> data's fill values.
#> Warning: No shared levels found between `names(values)` of the manual scale and the
#> data's fill values.
#> Warning: Removed 3 rows containing missing values or values outside the scale range
#> (`geom_path()`).
#> Warning: Removed 3 rows containing missing values or values outside the scale range
#> (`geom_path()`).
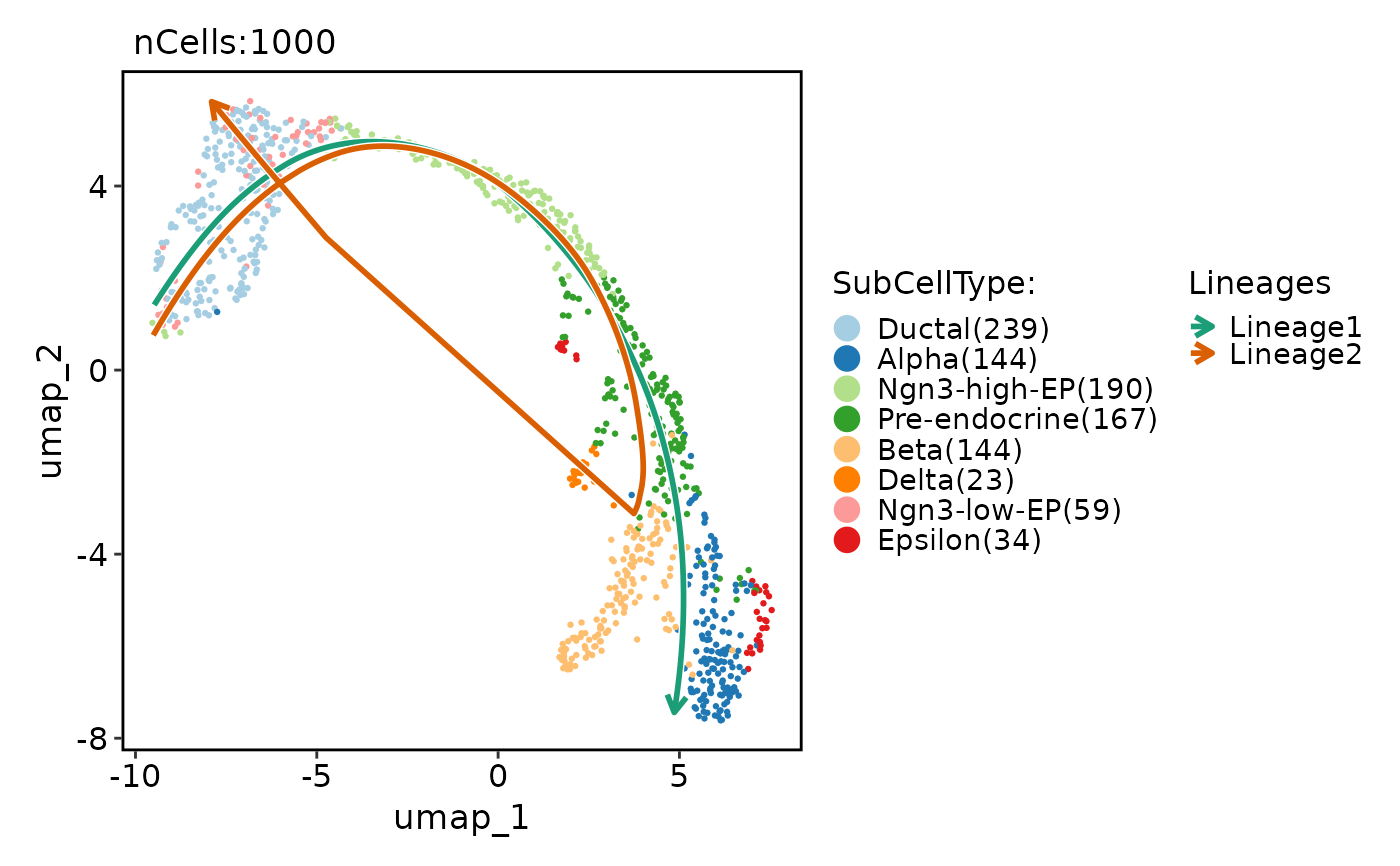 pancreas_sub <- RunDynamicFeatures(
pancreas_sub,
lineages = c("Lineage1", "Lineage2"),
n_candidates = 200
)
#> ℹ [2026-02-11 03:17:12] Start find dynamic features
#> ℹ [2026-02-11 03:17:15] Data type is raw counts
#> ℹ [2026-02-11 03:17:17] Number of candidate features (union): 231
#> ℹ [2026-02-11 03:17:17] Data type is raw counts
#> ℹ [2026-02-11 03:17:17] Calculating dynamic features for "Lineage1"...
#> ℹ [2026-02-11 03:17:17] Using 1 core
#> ⠙ [2026-02-11 03:17:17] Running for Gcg [1/231] ■ …
#> ⠹ [2026-02-11 03:17:17] Running for Gm8773 [87/231] ■■■■■■■■■■■■ …
#> ⠸ [2026-02-11 03:17:17] Running for Tm4sf4 [199/231] ■■■■■■■■■■■■■■■■■■■■■■■■■■…
#> ✔ [2026-02-11 03:17:17] Completed 231 tasks in 6.3s
#>
#> ℹ [2026-02-11 03:17:17] Building results
#> ℹ [2026-02-11 03:17:23] Calculating dynamic features for "Lineage2"...
#> ℹ [2026-02-11 03:17:23] Using 1 core
#> ⠙ [2026-02-11 03:17:23] Running for Tuba1c [68/231] ■■■■■■■■■■ …
#> ⠹ [2026-02-11 03:17:23] Running for Sox9 [171/231] ■■■■■■■■■■■■■■■■■■■■■■■ …
#> ✔ [2026-02-11 03:17:23] Completed 231 tasks in 6.6s
#>
#> ℹ [2026-02-11 03:17:23] Building results
#> ✔ [2026-02-11 03:17:30] Find dynamic features done
ht1 <- DynamicHeatmap(
pancreas_sub,
lineages = "Lineage1",
n_split = 5,
split_method = "kmeans-peaktime",
cell_annotation = "SubCellType"
)
#> ℹ [2026-02-11 03:18:04] [1] 162 features from Lineage1 passed the threshold (exp_ncells>[1] 20 & r.sq>[1] 0.2 & dev.expl>[1] 0.2 & padjust<[1] 0.05):
#> ℹ Gcg,Ghrl,Iapp,Pyy,Rbp4,Lrpprc,Slc38a5,Cdkn1a,2810417H13Rik,Chga...
#> ℹ [2026-02-11 03:18:05]
#> ℹ The size of the heatmap is fixed because certain elements are not scalable.
#> ℹ The width and height of the heatmap are determined by the size of the current viewport.
#> ℹ If you want to have more control over the size, you can manually set the parameters 'width' and 'height'.
pancreas_sub <- RunDynamicFeatures(
pancreas_sub,
lineages = c("Lineage1", "Lineage2"),
n_candidates = 200
)
#> ℹ [2026-02-11 03:17:12] Start find dynamic features
#> ℹ [2026-02-11 03:17:15] Data type is raw counts
#> ℹ [2026-02-11 03:17:17] Number of candidate features (union): 231
#> ℹ [2026-02-11 03:17:17] Data type is raw counts
#> ℹ [2026-02-11 03:17:17] Calculating dynamic features for "Lineage1"...
#> ℹ [2026-02-11 03:17:17] Using 1 core
#> ⠙ [2026-02-11 03:17:17] Running for Gcg [1/231] ■ …
#> ⠹ [2026-02-11 03:17:17] Running for Gm8773 [87/231] ■■■■■■■■■■■■ …
#> ⠸ [2026-02-11 03:17:17] Running for Tm4sf4 [199/231] ■■■■■■■■■■■■■■■■■■■■■■■■■■…
#> ✔ [2026-02-11 03:17:17] Completed 231 tasks in 6.3s
#>
#> ℹ [2026-02-11 03:17:17] Building results
#> ℹ [2026-02-11 03:17:23] Calculating dynamic features for "Lineage2"...
#> ℹ [2026-02-11 03:17:23] Using 1 core
#> ⠙ [2026-02-11 03:17:23] Running for Tuba1c [68/231] ■■■■■■■■■■ …
#> ⠹ [2026-02-11 03:17:23] Running for Sox9 [171/231] ■■■■■■■■■■■■■■■■■■■■■■■ …
#> ✔ [2026-02-11 03:17:23] Completed 231 tasks in 6.6s
#>
#> ℹ [2026-02-11 03:17:23] Building results
#> ✔ [2026-02-11 03:17:30] Find dynamic features done
ht1 <- DynamicHeatmap(
pancreas_sub,
lineages = "Lineage1",
n_split = 5,
split_method = "kmeans-peaktime",
cell_annotation = "SubCellType"
)
#> ℹ [2026-02-11 03:18:04] [1] 162 features from Lineage1 passed the threshold (exp_ncells>[1] 20 & r.sq>[1] 0.2 & dev.expl>[1] 0.2 & padjust<[1] 0.05):
#> ℹ Gcg,Ghrl,Iapp,Pyy,Rbp4,Lrpprc,Slc38a5,Cdkn1a,2810417H13Rik,Chga...
#> ℹ [2026-02-11 03:18:05]
#> ℹ The size of the heatmap is fixed because certain elements are not scalable.
#> ℹ The width and height of the heatmap are determined by the size of the current viewport.
#> ℹ If you want to have more control over the size, you can manually set the parameters 'width' and 'height'.
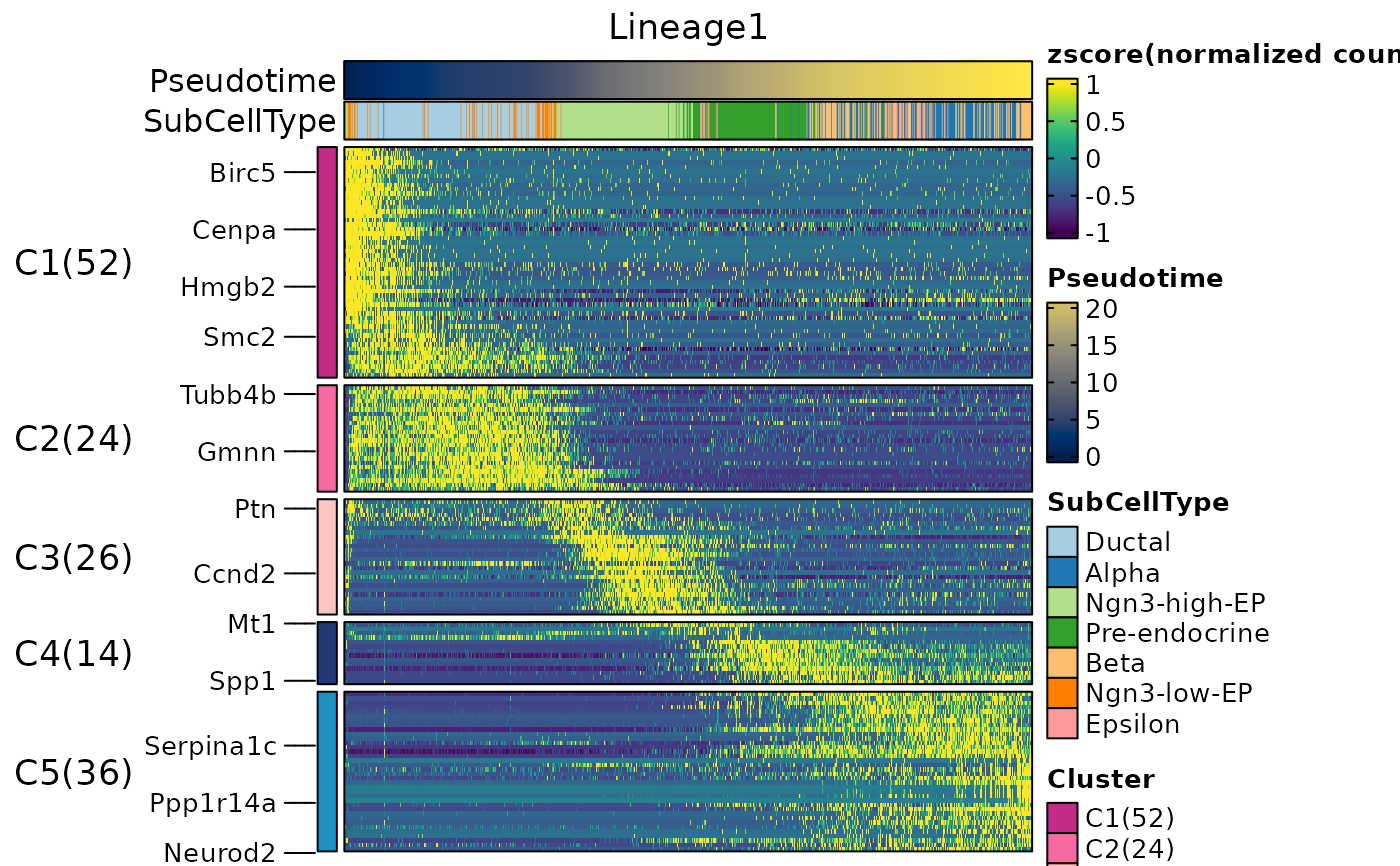 ht1$plot
ht1$plot
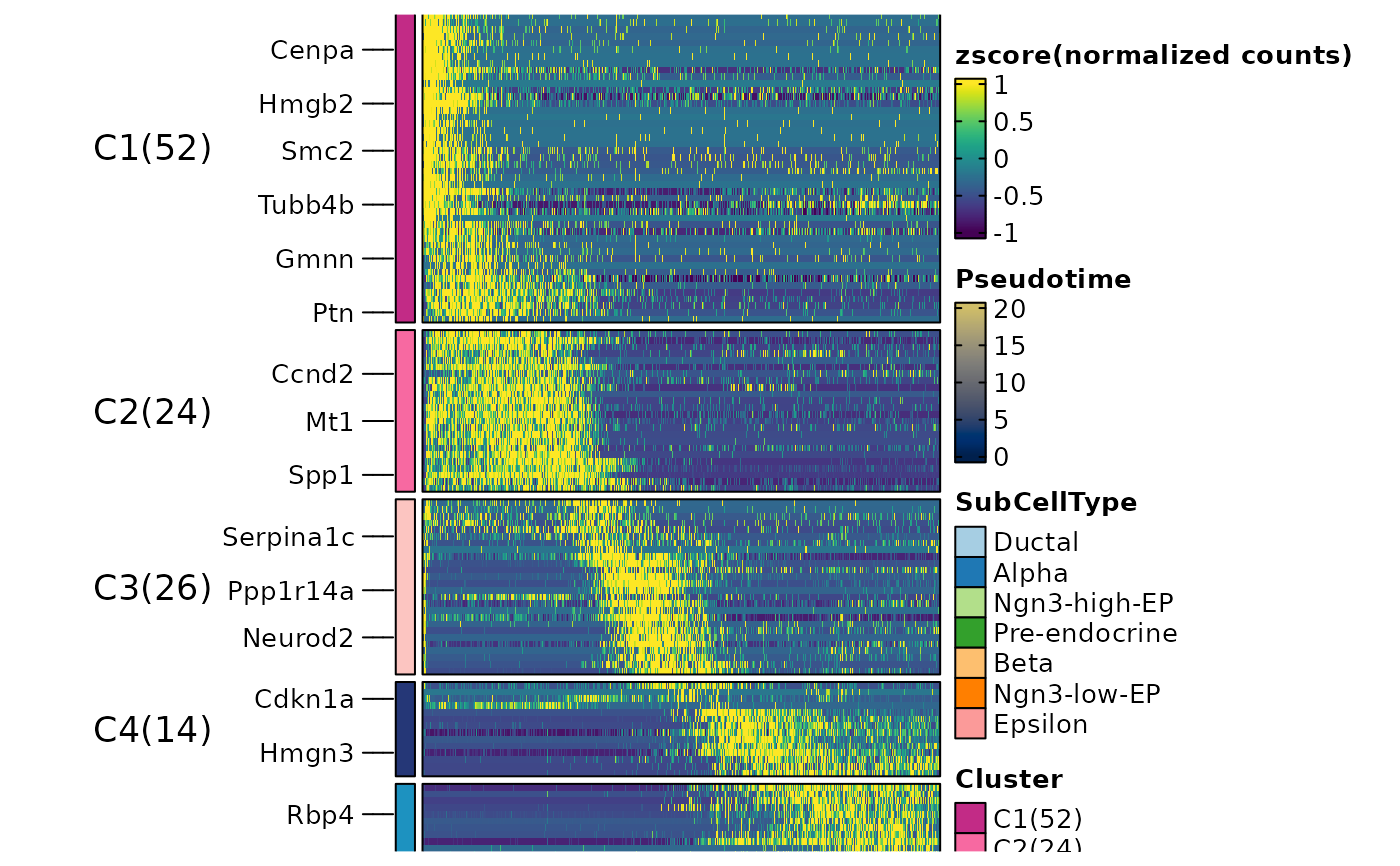 thisplot::panel_fix(ht1$plot, raster = TRUE, dpi = 50)
thisplot::panel_fix(ht1$plot, raster = TRUE, dpi = 50)
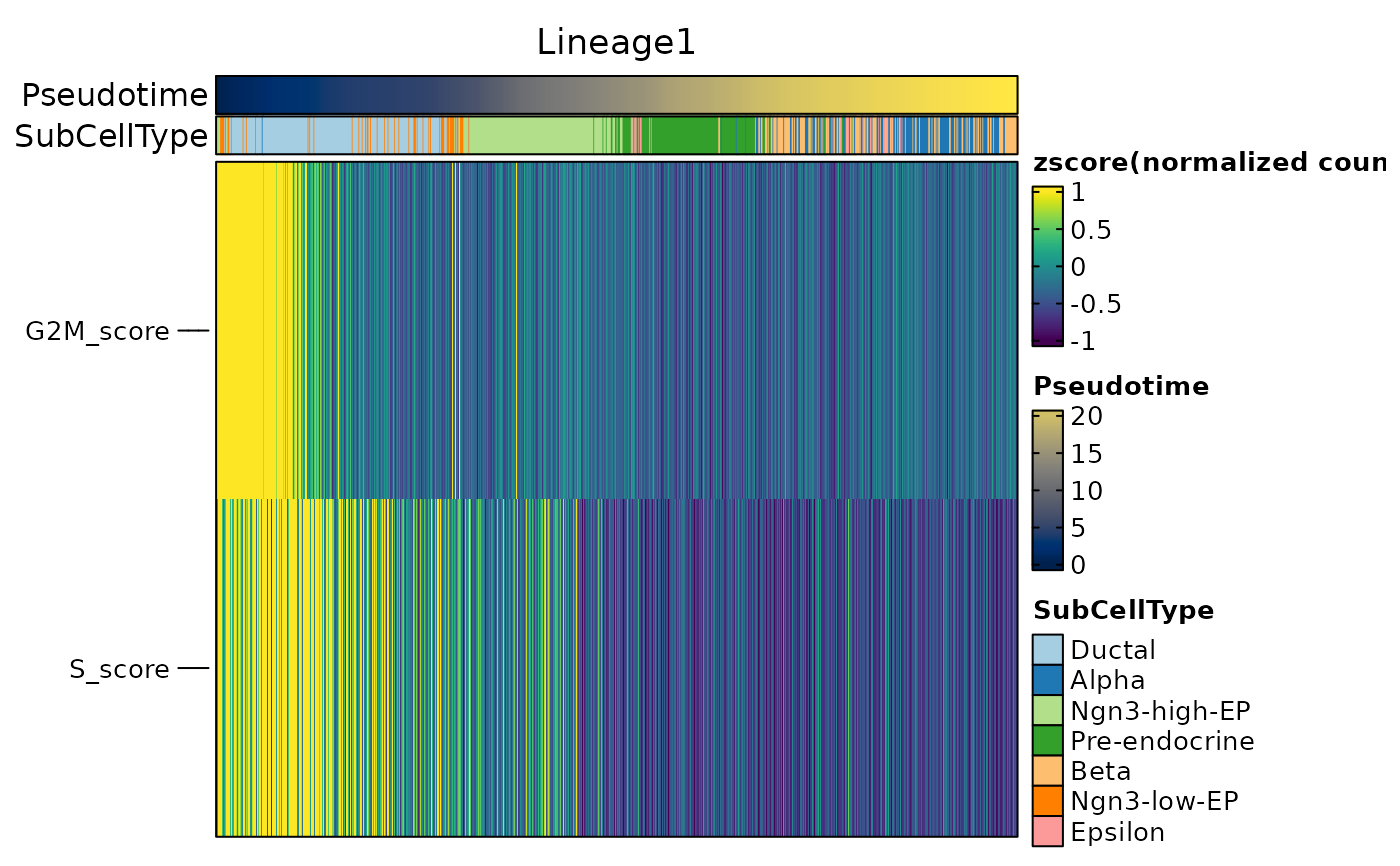
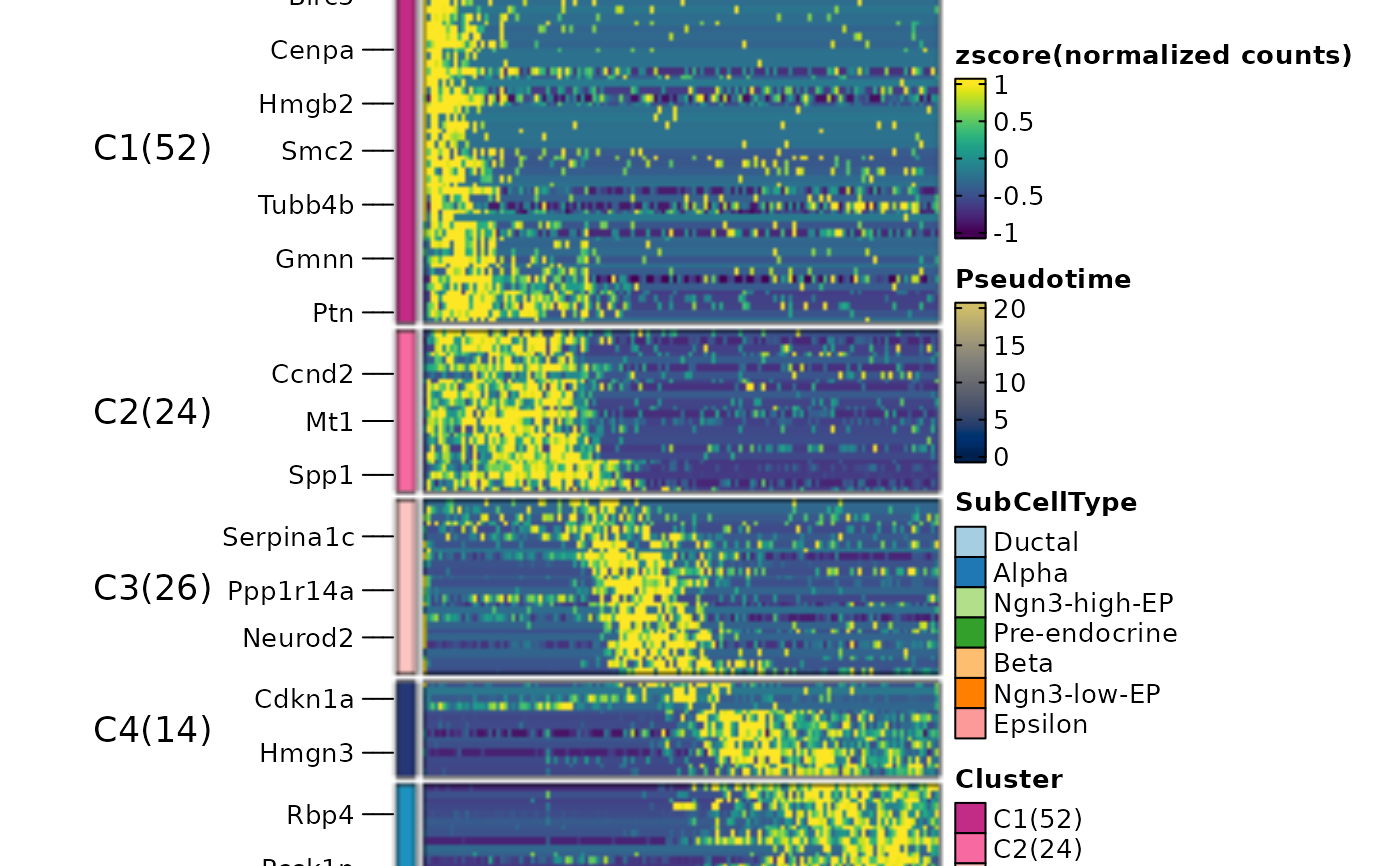 ht2 <- DynamicHeatmap(
pancreas_sub,
lineages = "Lineage1",
features = c(
"Sox9",
"Neurod2",
"Isl1",
"Rbp4",
"Pyy", "S_score", "G2M_score"
),
cell_annotation = "SubCellType"
)
#> ℹ [2026-02-11 03:18:07] Start find dynamic features
#> ℹ [2026-02-11 03:18:07] Data type is raw counts
#> ℹ [2026-02-11 03:18:08] Number of candidate features (union): 2
#> ℹ [2026-02-11 03:18:08] Data type is raw counts
#> ! [2026-02-11 03:18:08] Negative values detected
#> ! [2026-02-11 03:18:08] Negative values detected
#> ℹ [2026-02-11 03:18:08] Calculating dynamic features for "Lineage1"...
#> ℹ [2026-02-11 03:18:08] Using 1 core
#> ⠙ [2026-02-11 03:18:08] Running for S_score [1/2] ■■■■■■■■■■■■■■■■ …
#> ✔ [2026-02-11 03:18:08] Completed 2 tasks in 51ms
#>
#> ℹ [2026-02-11 03:18:08] Building results
#> ✔ [2026-02-11 03:18:08] Find dynamic features done
#> ℹ [2026-02-11 03:18:08] Some features were missing in at least one lineage:
#> ℹ Isl1,Neurod2,Pyy,Rbp4,Sox9...
ht2$plot
ht2 <- DynamicHeatmap(
pancreas_sub,
lineages = "Lineage1",
features = c(
"Sox9",
"Neurod2",
"Isl1",
"Rbp4",
"Pyy", "S_score", "G2M_score"
),
cell_annotation = "SubCellType"
)
#> ℹ [2026-02-11 03:18:07] Start find dynamic features
#> ℹ [2026-02-11 03:18:07] Data type is raw counts
#> ℹ [2026-02-11 03:18:08] Number of candidate features (union): 2
#> ℹ [2026-02-11 03:18:08] Data type is raw counts
#> ! [2026-02-11 03:18:08] Negative values detected
#> ! [2026-02-11 03:18:08] Negative values detected
#> ℹ [2026-02-11 03:18:08] Calculating dynamic features for "Lineage1"...
#> ℹ [2026-02-11 03:18:08] Using 1 core
#> ⠙ [2026-02-11 03:18:08] Running for S_score [1/2] ■■■■■■■■■■■■■■■■ …
#> ✔ [2026-02-11 03:18:08] Completed 2 tasks in 51ms
#>
#> ℹ [2026-02-11 03:18:08] Building results
#> ✔ [2026-02-11 03:18:08] Find dynamic features done
#> ℹ [2026-02-11 03:18:08] Some features were missing in at least one lineage:
#> ℹ Isl1,Neurod2,Pyy,Rbp4,Sox9...
ht2$plot
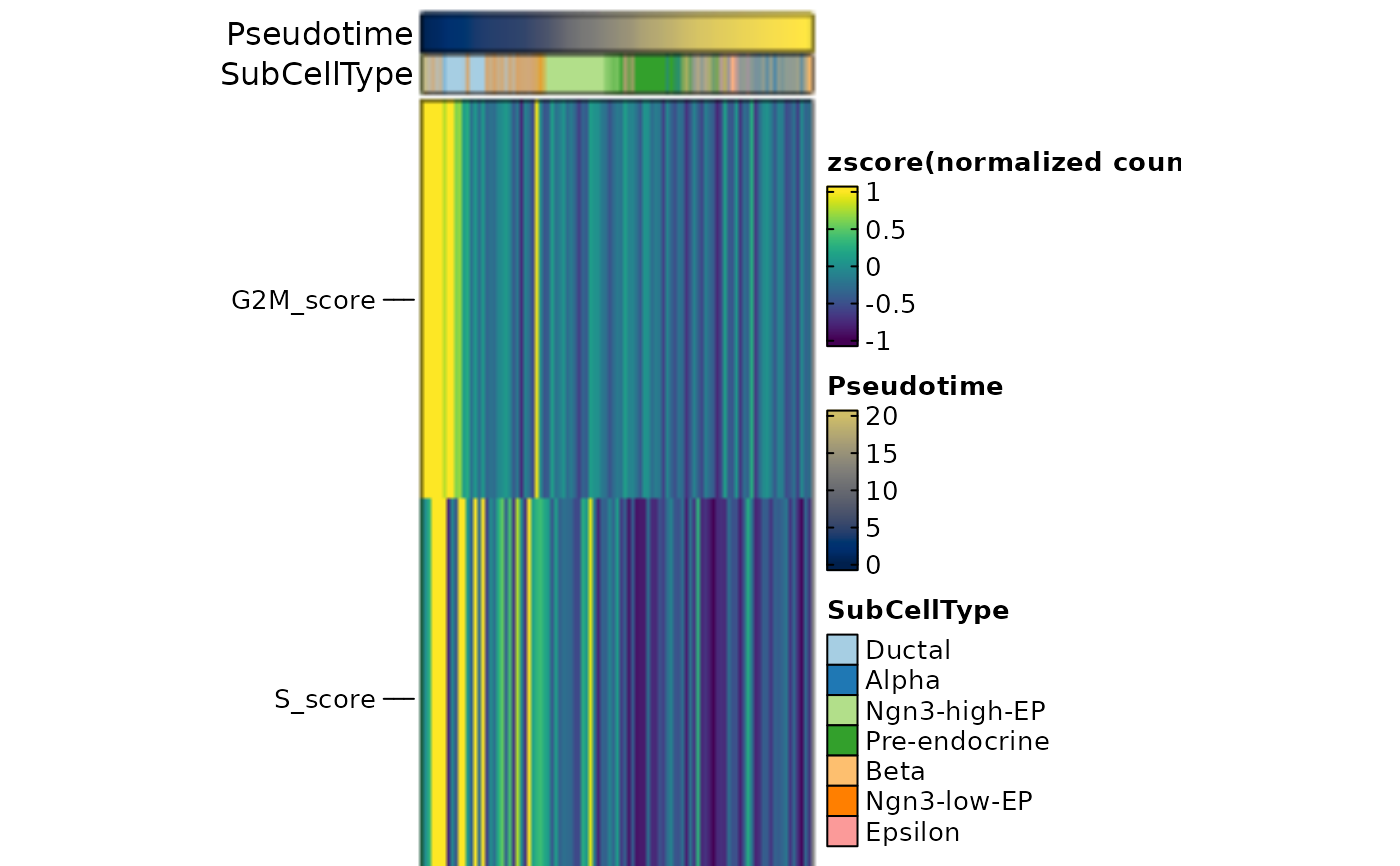
 thisplot::panel_fix(
ht2$plot,
height = 5,
width = 5,
raster = TRUE,
dpi = 50
)
thisplot::panel_fix(
ht2$plot,
height = 5,
width = 5,
raster = TRUE,
dpi = 50
)
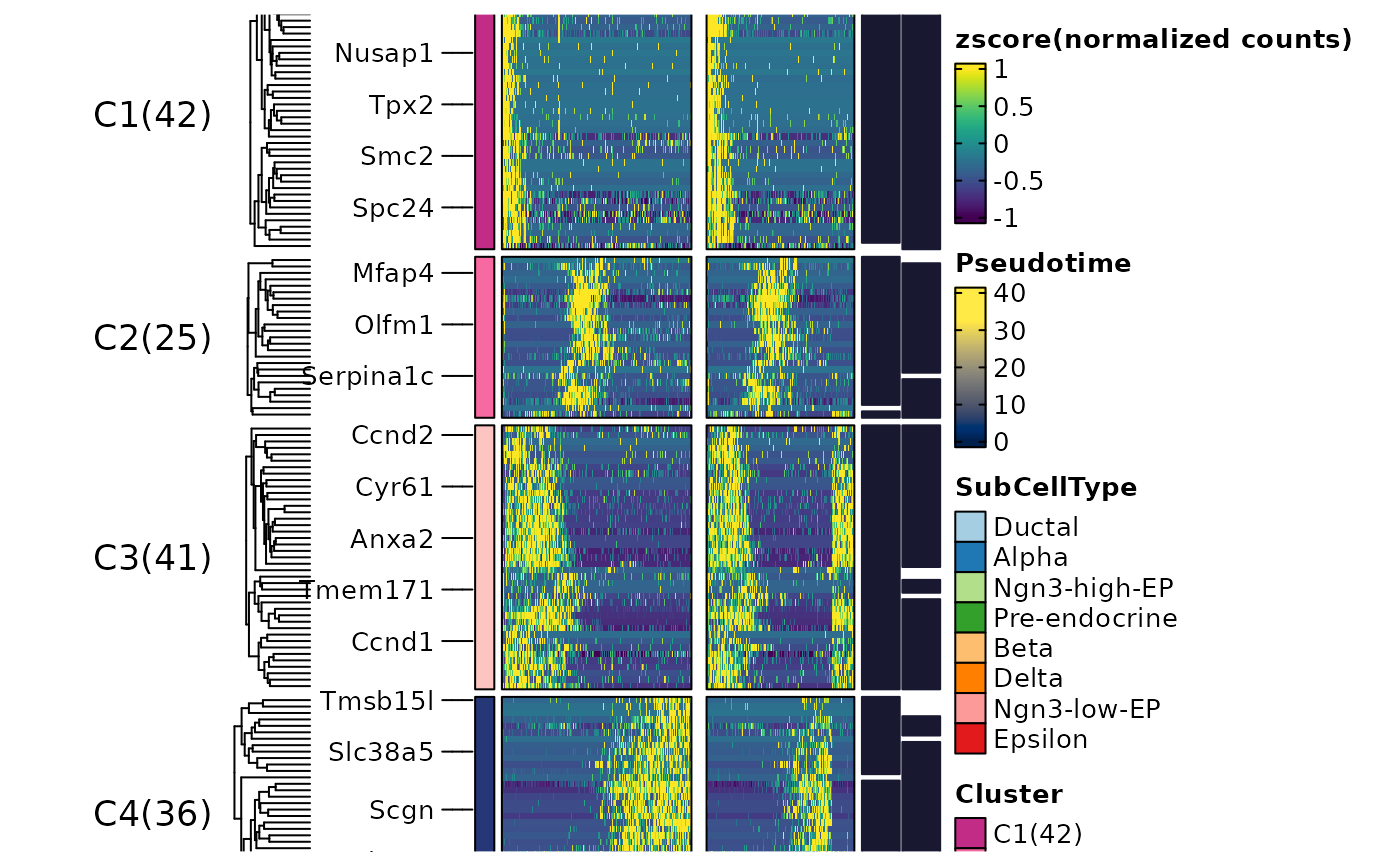
 ht3 <- DynamicHeatmap(
pancreas_sub,
lineages = c("Lineage1", "Lineage2"),
n_split = 5,
split_method = "kmeans",
cluster_rows = TRUE,
cell_annotation = "SubCellType"
)
#> ℹ [2026-02-11 03:18:10] [1] 180 features from Lineage1,Lineage2 passed the threshold (exp_ncells>[1] 20 & r.sq>[1] 0.2 & dev.expl>[1] 0.2 & padjust<[1] 0.05):
#> ℹ Gcg,Ghrl,Iapp,Pyy,Rbp4,Lrpprc,Slc38a5,Cdkn1a,2810417H13Rik,Chga...
#> ℹ [2026-02-11 03:18:11]
#> ℹ The size of the heatmap is fixed because certain elements are not scalable.
#> ℹ The width and height of the heatmap are determined by the size of the current viewport.
#> ℹ If you want to have more control over the size, you can manually set the parameters 'width' and 'height'.
ht3 <- DynamicHeatmap(
pancreas_sub,
lineages = c("Lineage1", "Lineage2"),
n_split = 5,
split_method = "kmeans",
cluster_rows = TRUE,
cell_annotation = "SubCellType"
)
#> ℹ [2026-02-11 03:18:10] [1] 180 features from Lineage1,Lineage2 passed the threshold (exp_ncells>[1] 20 & r.sq>[1] 0.2 & dev.expl>[1] 0.2 & padjust<[1] 0.05):
#> ℹ Gcg,Ghrl,Iapp,Pyy,Rbp4,Lrpprc,Slc38a5,Cdkn1a,2810417H13Rik,Chga...
#> ℹ [2026-02-11 03:18:11]
#> ℹ The size of the heatmap is fixed because certain elements are not scalable.
#> ℹ The width and height of the heatmap are determined by the size of the current viewport.
#> ℹ If you want to have more control over the size, you can manually set the parameters 'width' and 'height'.
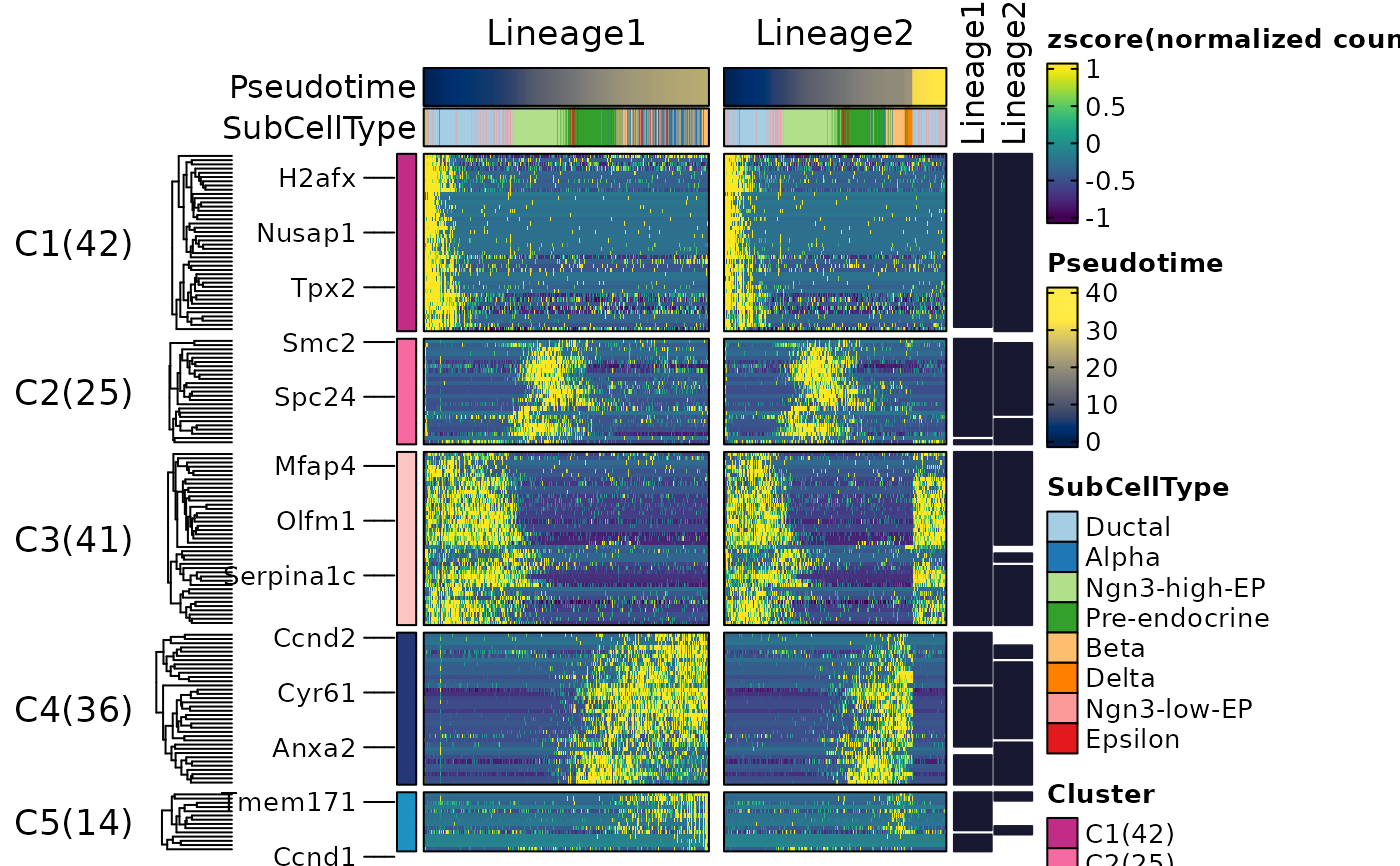 ht3$plot
ht3$plot
 ht4 <- DynamicHeatmap(
pancreas_sub,
lineages = c("Lineage1", "Lineage2"),
reverse_ht = "Lineage1",
cell_annotation = "SubCellType",
n_split = 5,
split_method = "mfuzz",
species = "Mus_musculus",
db = "GO_BP",
anno_terms = TRUE,
anno_keys = TRUE,
anno_features = TRUE
)
#> ℹ [2026-02-11 03:18:14] [1] 180 features from Lineage1,Lineage2 passed the threshold (exp_ncells>[1] 20 & r.sq>[1] 0.2 & dev.expl>[1] 0.2 & padjust<[1] 0.05):
#> ℹ Gcg,Ghrl,Iapp,Pyy,Rbp4,Lrpprc,Slc38a5,Cdkn1a,2810417H13Rik,Chga...
#> ℹ [2026-02-11 03:18:15] Start Enrichment analysis
#> ℹ [2026-02-11 03:22:52] Species: "Mus_musculus"
#> ℹ [2026-02-11 03:27:15] Preparing database: GO_BP
#> ℹ [2026-02-11 03:27:36] Convert ID types for the GO_BP database
#> ℹ [2026-02-11 03:27:36] Connect to the Ensembl archives...
#> ℹ [2026-02-11 03:27:36] Using the 115 version of ensembl database...
#> ℹ [2026-02-11 03:27:36] Downloading the ensembl database from https://sep2025.archive.ensembl.org...
#> ℹ [2026-02-11 03:27:38] Searching the dataset mmusculus ...
#> ℹ [2026-02-11 03:27:38] Connecting to the dataset mmusculus_gene_ensembl ...
#> ℹ [2026-02-11 03:27:39] Converting the geneIDs...
#> ℹ [2026-02-11 03:27:45] 23214 genes mapped with "entrez_id"
#> ℹ [2026-02-11 03:27:45] ==============================
#> ℹ 23214 genes mapped
#> ℹ 2516 genes unmapped
#> ℹ ==============================
#> ℹ [2026-02-11 03:27:59] Permform enrichment...
#> ℹ [2026-02-11 03:27:59] Using 1 core
#> ⠙ [2026-02-11 03:27:59] Running for 1 [1/5] ■■■■■■■ 2…
#> ⠹ [2026-02-11 03:27:59] Running for 2 [2/5] ■■■■■■■■■■■■■ 4…
#> ⠸ [2026-02-11 03:27:59] Running for 3 [3/5] ■■■■■■■■■■■■■■■■■■■ 6…
#> ⠼ [2026-02-11 03:27:59] Running for 4 [4/5] ■■■■■■■■■■■■■■■■■■■■■■■■■ 8…
#> ✔ [2026-02-11 03:27:59] Completed 5 tasks in 30.5s
#>
#> ℹ [2026-02-11 03:27:59] Building results
#> ✔ [2026-02-11 03:28:30] Enrichment analysis done
#> ℹ [2026-02-11 03:30:37]
#> ℹ The size of the heatmap is fixed because certain elements are not scalable.
#> ℹ The width and height of the heatmap are determined by the size of the current viewport.
#> ℹ If you want to have more control over the size, you can manually set the parameters 'width' and 'height'.
ht4 <- DynamicHeatmap(
pancreas_sub,
lineages = c("Lineage1", "Lineage2"),
reverse_ht = "Lineage1",
cell_annotation = "SubCellType",
n_split = 5,
split_method = "mfuzz",
species = "Mus_musculus",
db = "GO_BP",
anno_terms = TRUE,
anno_keys = TRUE,
anno_features = TRUE
)
#> ℹ [2026-02-11 03:18:14] [1] 180 features from Lineage1,Lineage2 passed the threshold (exp_ncells>[1] 20 & r.sq>[1] 0.2 & dev.expl>[1] 0.2 & padjust<[1] 0.05):
#> ℹ Gcg,Ghrl,Iapp,Pyy,Rbp4,Lrpprc,Slc38a5,Cdkn1a,2810417H13Rik,Chga...
#> ℹ [2026-02-11 03:18:15] Start Enrichment analysis
#> ℹ [2026-02-11 03:22:52] Species: "Mus_musculus"
#> ℹ [2026-02-11 03:27:15] Preparing database: GO_BP
#> ℹ [2026-02-11 03:27:36] Convert ID types for the GO_BP database
#> ℹ [2026-02-11 03:27:36] Connect to the Ensembl archives...
#> ℹ [2026-02-11 03:27:36] Using the 115 version of ensembl database...
#> ℹ [2026-02-11 03:27:36] Downloading the ensembl database from https://sep2025.archive.ensembl.org...
#> ℹ [2026-02-11 03:27:38] Searching the dataset mmusculus ...
#> ℹ [2026-02-11 03:27:38] Connecting to the dataset mmusculus_gene_ensembl ...
#> ℹ [2026-02-11 03:27:39] Converting the geneIDs...
#> ℹ [2026-02-11 03:27:45] 23214 genes mapped with "entrez_id"
#> ℹ [2026-02-11 03:27:45] ==============================
#> ℹ 23214 genes mapped
#> ℹ 2516 genes unmapped
#> ℹ ==============================
#> ℹ [2026-02-11 03:27:59] Permform enrichment...
#> ℹ [2026-02-11 03:27:59] Using 1 core
#> ⠙ [2026-02-11 03:27:59] Running for 1 [1/5] ■■■■■■■ 2…
#> ⠹ [2026-02-11 03:27:59] Running for 2 [2/5] ■■■■■■■■■■■■■ 4…
#> ⠸ [2026-02-11 03:27:59] Running for 3 [3/5] ■■■■■■■■■■■■■■■■■■■ 6…
#> ⠼ [2026-02-11 03:27:59] Running for 4 [4/5] ■■■■■■■■■■■■■■■■■■■■■■■■■ 8…
#> ✔ [2026-02-11 03:27:59] Completed 5 tasks in 30.5s
#>
#> ℹ [2026-02-11 03:27:59] Building results
#> ✔ [2026-02-11 03:28:30] Enrichment analysis done
#> ℹ [2026-02-11 03:30:37]
#> ℹ The size of the heatmap is fixed because certain elements are not scalable.
#> ℹ The width and height of the heatmap are determined by the size of the current viewport.
#> ℹ If you want to have more control over the size, you can manually set the parameters 'width' and 'height'.
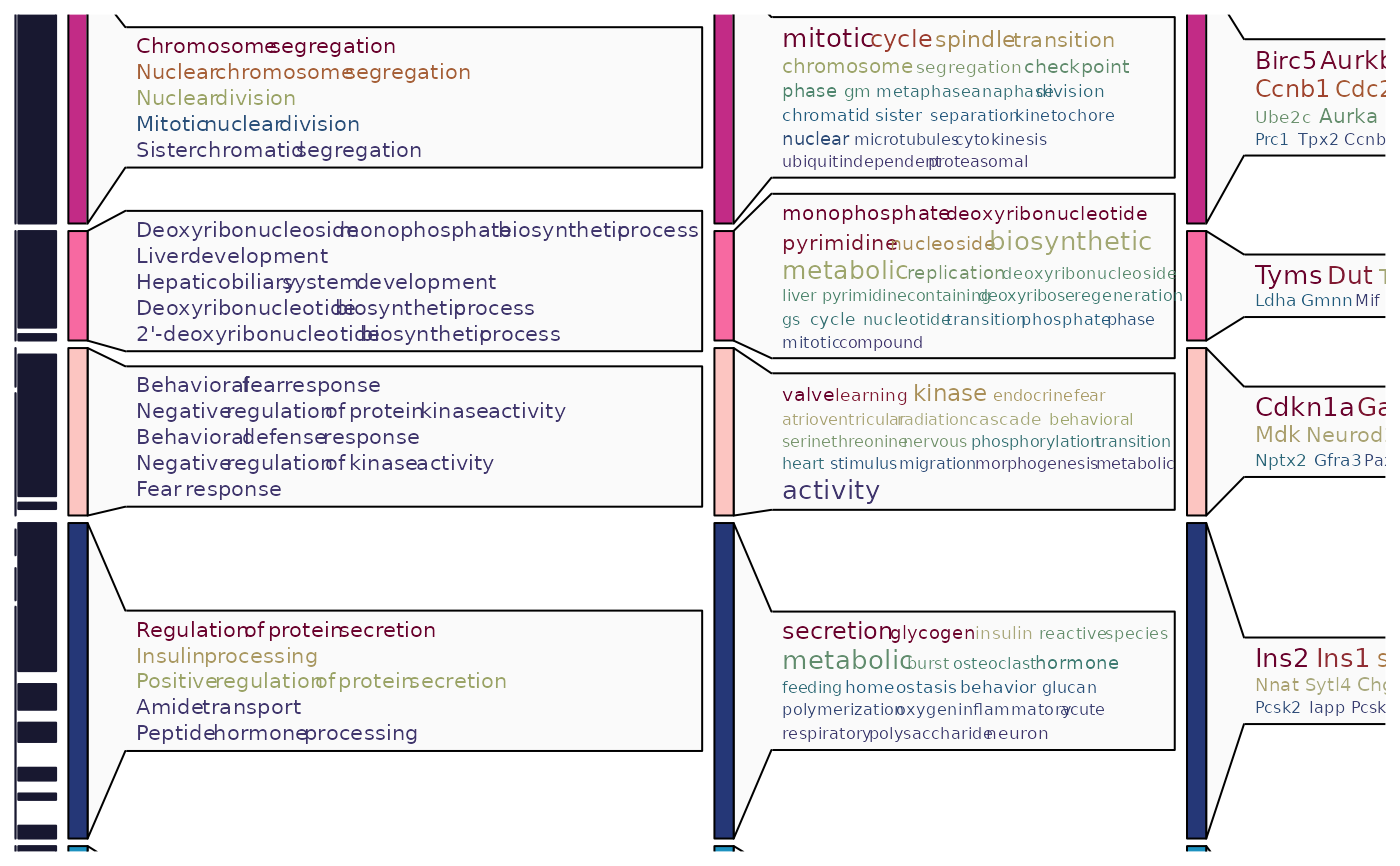
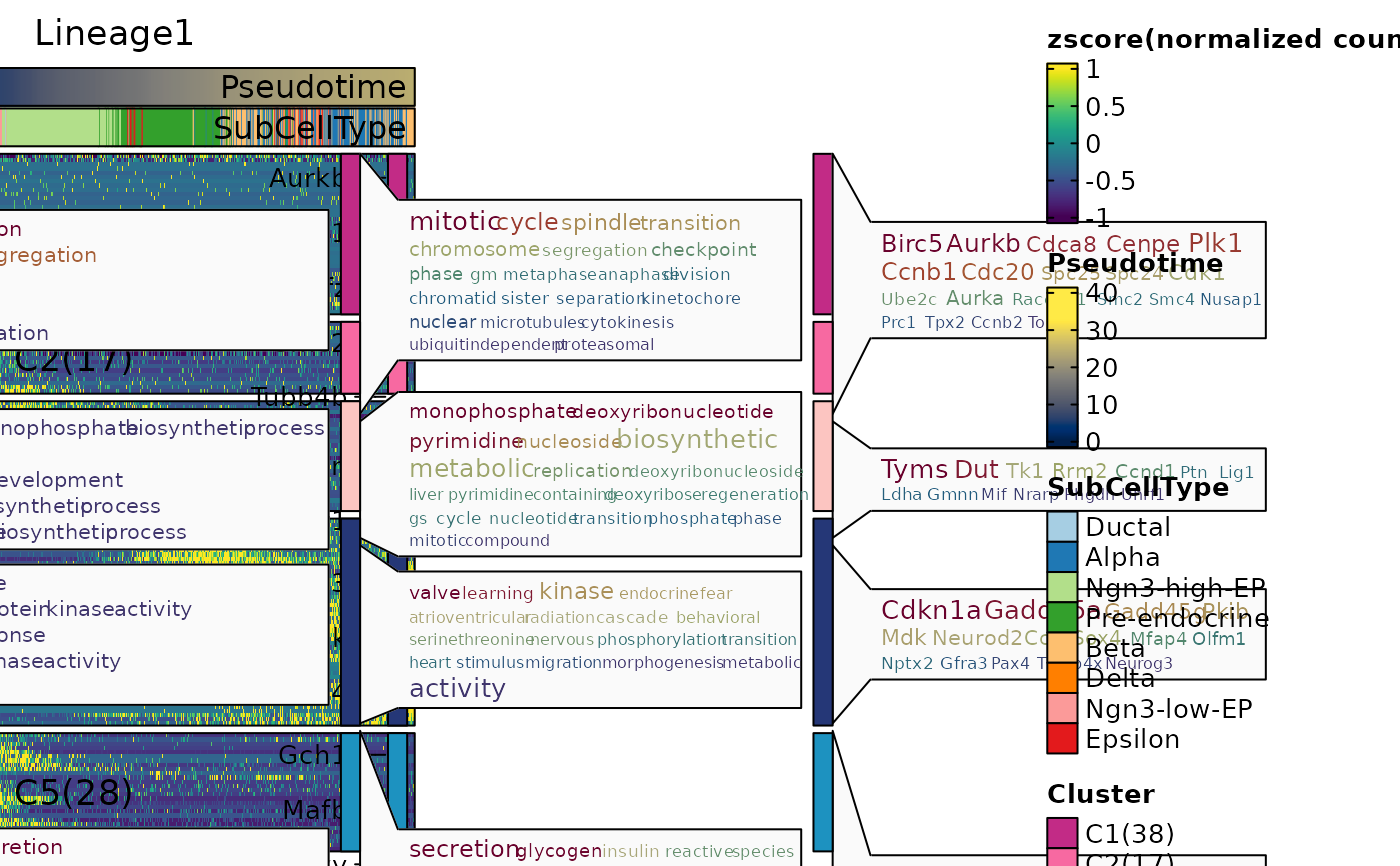 ht4$plot
ht4$plot
 if (FALSE) { # \dontrun{
pancreas_sub <- AnnotateFeatures(
pancreas_sub,
species = "Mus_musculus",
db = c("CSPA", "TF")
)
ht5 <- DynamicHeatmap(
pancreas_sub,
lineages = c("Lineage1", "Lineage2"),
reverse_ht = "Lineage1",
use_fitted = TRUE,
n_split = 6,
split_method = "mfuzz",
heatmap_palette = "viridis",
cell_annotation = c(
"SubCellType", "Phase", "G2M_score"
),
cell_annotation_palette = c(
"Paired", "simspec", "Purples"
),
separate_annotation = list(
"SubCellType", c("Arxes1", "Ncoa2")
),
separate_annotation_palette = c(
"Paired", "Set1"
),
separate_annotation_params = list(
height = grid::unit(10, "mm")
),
feature_annotation = c("TF", "CSPA"),
feature_annotation_palcolor = list(
c("gold", "steelblue"),
c("forestgreen")
),
pseudotime_label = 25,
pseudotime_label_color = "red"
)
ht5$plot
ht6 <- DynamicHeatmap(
pancreas_sub,
lineages = c("Lineage1", "Lineage2"),
reverse_ht = "Lineage1",
use_fitted = TRUE,
n_split = 6,
split_method = "mfuzz",
heatmap_palette = "viridis",
cell_annotation = c(
"SubCellType", "Phase", "G2M_score"
),
cell_annotation_palette = c(
"Paired", "simspec", "Purples"
),
separate_annotation = list(
"SubCellType", c("Arxes1", "Ncoa2")
),
separate_annotation_palette = c("Paired", "Set1"),
separate_annotation_params = list(width = grid::unit(10, "mm")),
feature_annotation = c("TF", "CSPA"),
feature_annotation_palcolor = list(
c("gold", "steelblue"),
c("forestgreen")
),
pseudotime_label = 25,
pseudotime_label_color = "red",
flip = TRUE, column_title_rot = 45
)
ht6$plot
} # }
if (FALSE) { # \dontrun{
pancreas_sub <- AnnotateFeatures(
pancreas_sub,
species = "Mus_musculus",
db = c("CSPA", "TF")
)
ht5 <- DynamicHeatmap(
pancreas_sub,
lineages = c("Lineage1", "Lineage2"),
reverse_ht = "Lineage1",
use_fitted = TRUE,
n_split = 6,
split_method = "mfuzz",
heatmap_palette = "viridis",
cell_annotation = c(
"SubCellType", "Phase", "G2M_score"
),
cell_annotation_palette = c(
"Paired", "simspec", "Purples"
),
separate_annotation = list(
"SubCellType", c("Arxes1", "Ncoa2")
),
separate_annotation_palette = c(
"Paired", "Set1"
),
separate_annotation_params = list(
height = grid::unit(10, "mm")
),
feature_annotation = c("TF", "CSPA"),
feature_annotation_palcolor = list(
c("gold", "steelblue"),
c("forestgreen")
),
pseudotime_label = 25,
pseudotime_label_color = "red"
)
ht5$plot
ht6 <- DynamicHeatmap(
pancreas_sub,
lineages = c("Lineage1", "Lineage2"),
reverse_ht = "Lineage1",
use_fitted = TRUE,
n_split = 6,
split_method = "mfuzz",
heatmap_palette = "viridis",
cell_annotation = c(
"SubCellType", "Phase", "G2M_score"
),
cell_annotation_palette = c(
"Paired", "simspec", "Purples"
),
separate_annotation = list(
"SubCellType", c("Arxes1", "Ncoa2")
),
separate_annotation_palette = c("Paired", "Set1"),
separate_annotation_params = list(width = grid::unit(10, "mm")),
feature_annotation = c("TF", "CSPA"),
feature_annotation_palcolor = list(
c("gold", "steelblue"),
c("forestgreen")
),
pseudotime_label = 25,
pseudotime_label_color = "red",
flip = TRUE, column_title_rot = 45
)
ht6$plot
} # }