Runs the Slingshot algorithm on a Seurat object.
Usage
RunSlingshot(
srt,
group.by,
reduction = NULL,
dims = NULL,
start = NULL,
end = NULL,
prefix = NULL,
reverse = FALSE,
align_start = FALSE,
show_plot = TRUE,
lineage_palette = "Dark2",
seed = 11,
...
)Arguments
- srt
A Seurat object.
- group.by
Name of one or more meta.data columns to group (color) cells by.
- reduction
Which dimensionality reduction to use. If not specified, will use the reduction returned by DefaultReduction.
- dims
The dimensions to use for the Slingshot algorithm. Default is
NULL, which uses first two dimensions.- start
The starting group for the Slingshot algorithm. Default is
NULL.- end
The ending group for the Slingshot algorithm. Default is
NULL.- prefix
The prefix to add to the column names of the resulting pseudotime variable. Default is
NULL.- reverse
Logical value indicating whether to reverse the pseudotime variable. Default is
FALSE.- align_start
Logical value indicating whether to align the starting pseudotime values at the maximum pseudotime. Default is
FALSE.- show_plot
Logical value indicating whether to show the dimensionality plot. Default is
TRUE.- lineage_palette
The color palette to use for the lineages in the plot. Default is
"Dark2".- seed
Random seed for reproducibility. Default is
11.- ...
Additional arguments to be passed to the slingshot::slingshot function.
Examples
data(pancreas_sub)
pancreas_sub <- standard_scop(pancreas_sub)
#> ℹ [2026-02-11 04:17:59] Start standard scop workflow...
#> ℹ [2026-02-11 04:18:00] Checking a list of <Seurat>...
#> ! [2026-02-11 04:18:00] Data 1/1 of the `srt_list` is "unknown"
#> ℹ [2026-02-11 04:18:00] Perform `NormalizeData()` with `normalization.method = 'LogNormalize'` on the data 1/1 of the `srt_list`...
#> ℹ [2026-02-11 04:18:02] Perform `Seurat::FindVariableFeatures()` on the data 1/1 of the `srt_list`...
#> ℹ [2026-02-11 04:18:03] Use the separate HVF from srt_list
#> ℹ [2026-02-11 04:18:03] Number of available HVF: 2000
#> ℹ [2026-02-11 04:18:03] Finished check
#> ℹ [2026-02-11 04:18:03] Perform `Seurat::ScaleData()`
#> ℹ [2026-02-11 04:18:04] Perform pca linear dimension reduction
#> ℹ [2026-02-11 04:18:05] Perform `Seurat::FindClusters()` with `cluster_algorithm = 'louvain'` and `cluster_resolution = 0.6`
#> ℹ [2026-02-11 04:18:05] Reorder clusters...
#> ℹ [2026-02-11 04:18:05] Perform umap nonlinear dimension reduction
#> ℹ [2026-02-11 04:18:05] Non-linear dimensionality reduction (umap) using (Standardpca) dims (1-50) as input
#> ℹ [2026-02-11 04:18:09] Non-linear dimensionality reduction (umap) using (Standardpca) dims (1-50) as input
#> ✔ [2026-02-11 04:18:14] Run scop standard workflow completed
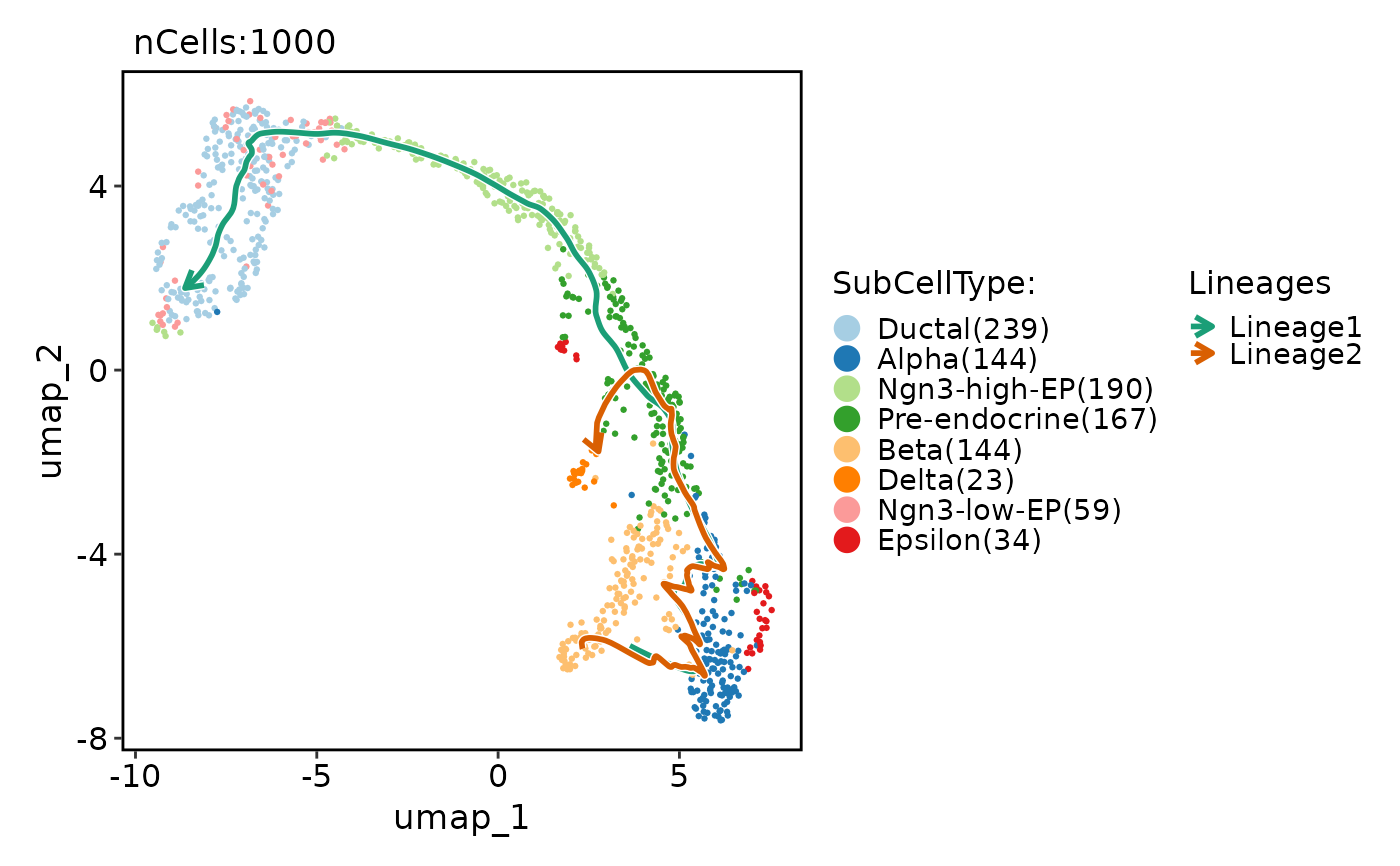
pancreas_sub <- RunSlingshot(
pancreas_sub,
group.by = "SubCellType",
reduction = "UMAP"
)
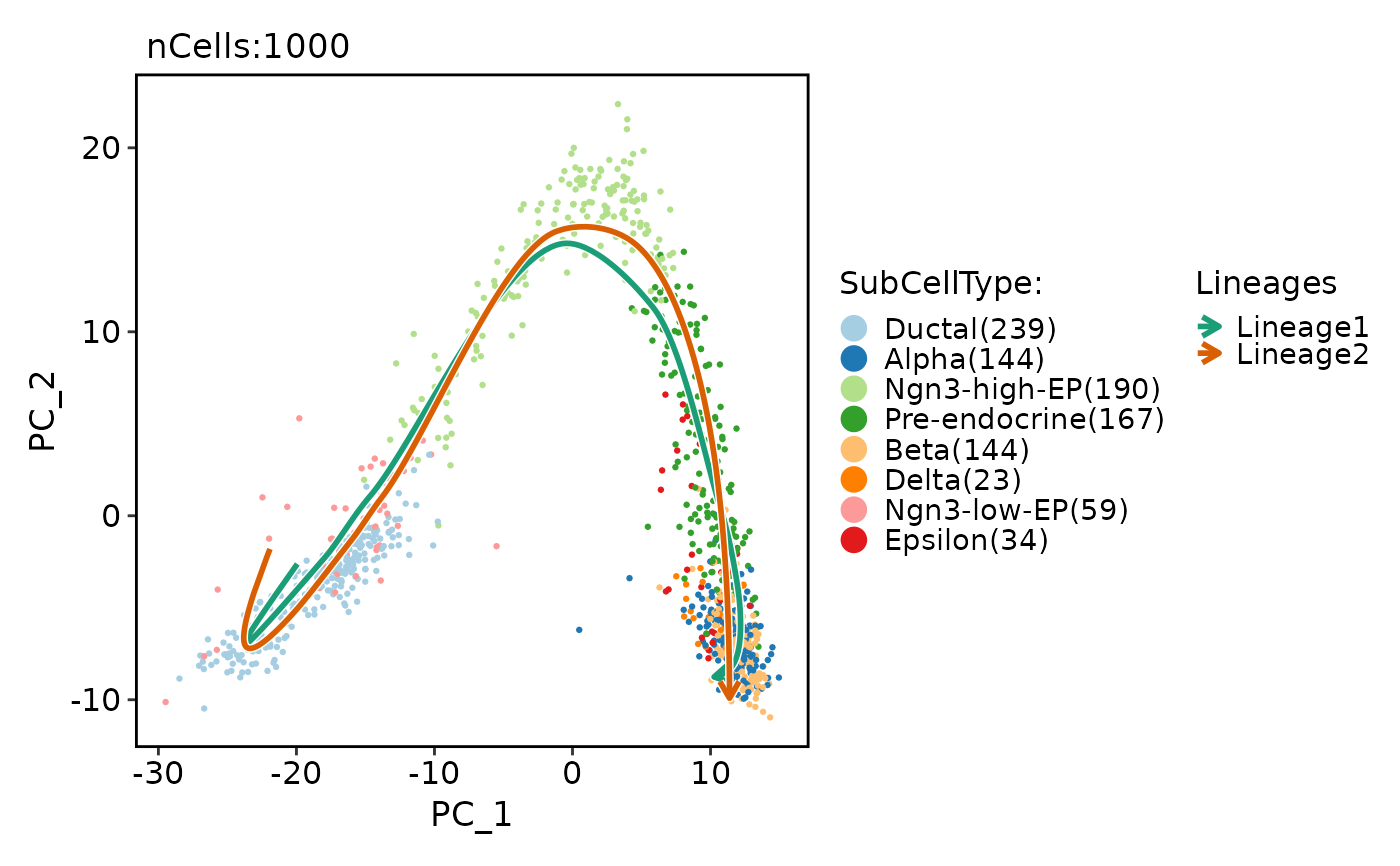
 pancreas_sub <- RunSlingshot(
pancreas_sub,
group.by = "SubCellType",
reduction = "PCA"
)
pancreas_sub <- RunSlingshot(
pancreas_sub,
group.by = "SubCellType",
reduction = "PCA"
)
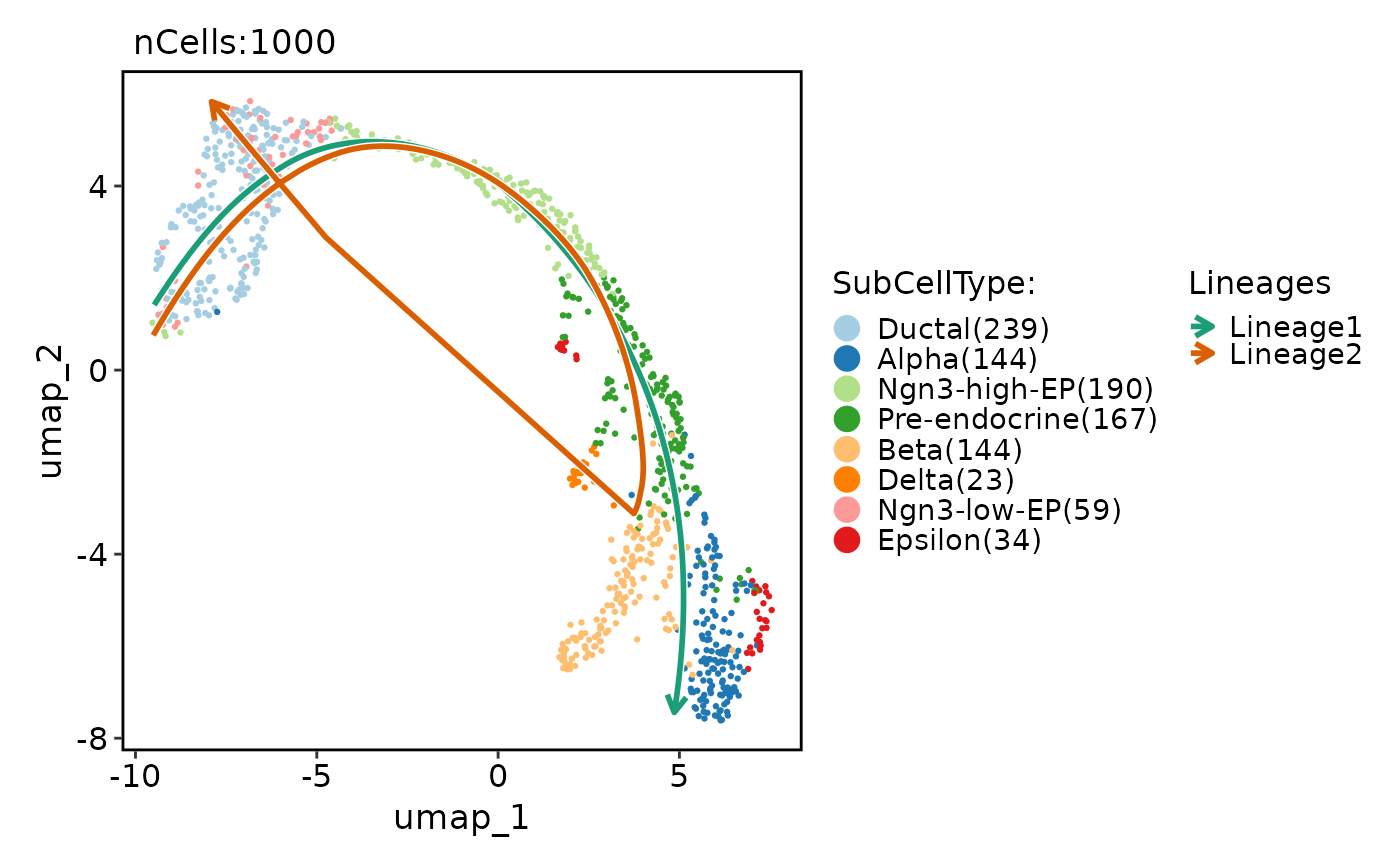
 CellDimPlot(
pancreas_sub,
group.by = "SubCellType",
reduction = "UMAP",
lineages = paste0("Lineage", 1:2),
lineages_span = 0.1
)
CellDimPlot(
pancreas_sub,
group.by = "SubCellType",
reduction = "UMAP",
lineages = paste0("Lineage", 1:2),
lineages_span = 0.1
)
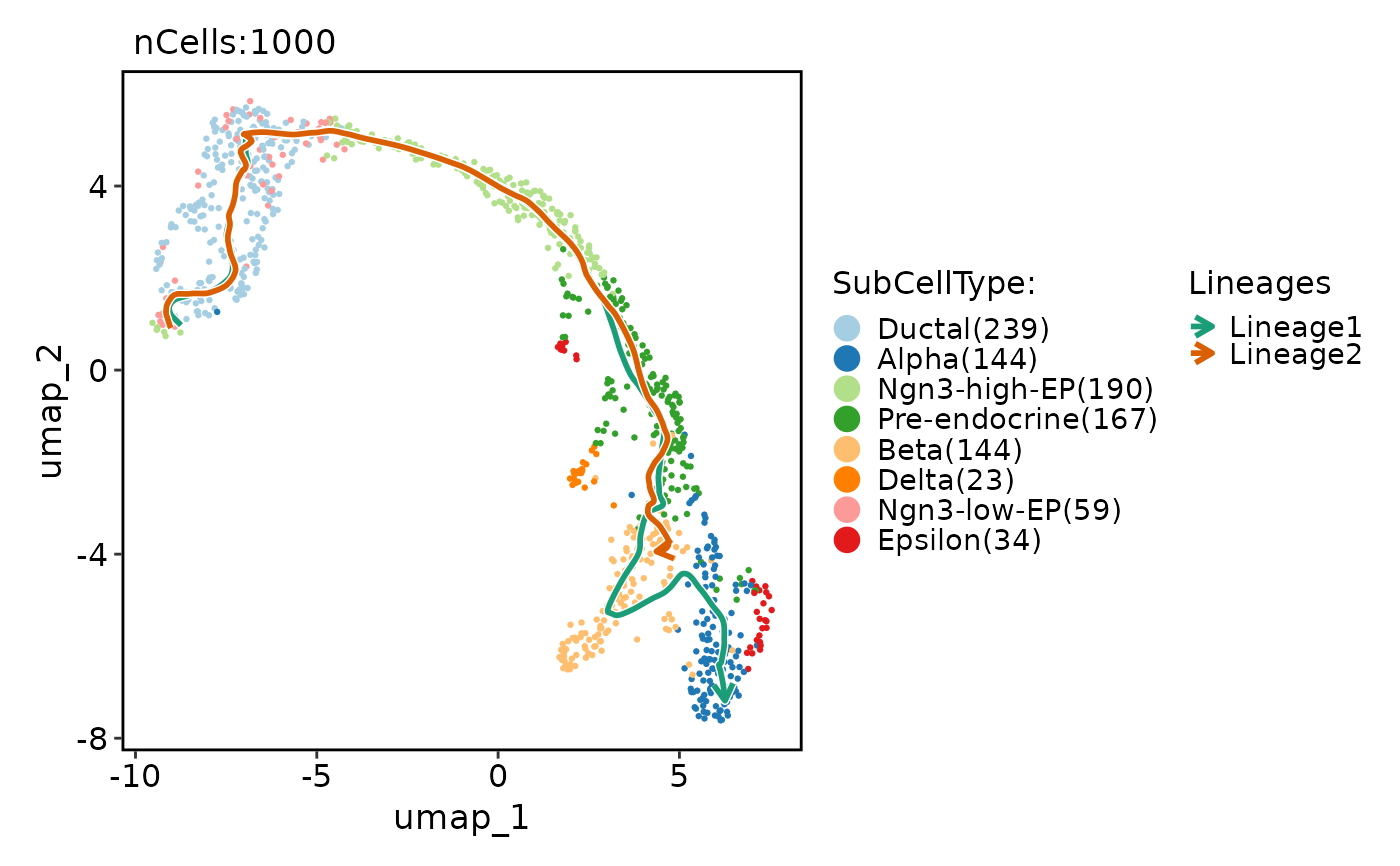 # 3D lineage
pancreas_sub <- RunSlingshot(
pancreas_sub,
group.by = "SubCellType",
reduction = "StandardpcaUMAP3D"
)
CellDimPlot(
pancreas_sub,
group.by = "SubCellType",
reduction = "UMAP",
lineages = paste0("Lineage", 1:2),
lineages_span = 0.1,
lineages_trim = c(0.05, 0.95)
)
# 3D lineage
pancreas_sub <- RunSlingshot(
pancreas_sub,
group.by = "SubCellType",
reduction = "StandardpcaUMAP3D"
)
CellDimPlot(
pancreas_sub,
group.by = "SubCellType",
reduction = "UMAP",
lineages = paste0("Lineage", 1:2),
lineages_span = 0.1,
lineages_trim = c(0.05, 0.95)
)