Integrate single-cell RNA-seq data using various integration methods.
Usage
integration_scop(
srt_merge = NULL,
batch,
append = TRUE,
srt_list = NULL,
assay = NULL,
integration_method = "Uncorrected",
do_normalization = NULL,
normalization_method = "LogNormalize",
do_HVF_finding = TRUE,
HVF_source = "separate",
HVF_method = "vst",
nHVF = 2000,
HVF_min_intersection = 1,
HVF = NULL,
do_scaling = TRUE,
vars_to_regress = NULL,
regression_model = "linear",
scale_within_batch = FALSE,
linear_reduction = "pca",
linear_reduction_dims = 50,
linear_reduction_dims_use = NULL,
linear_reduction_params = list(),
force_linear_reduction = FALSE,
nonlinear_reduction = "umap",
nonlinear_reduction_dims = c(2, 3),
nonlinear_reduction_params = list(),
force_nonlinear_reduction = TRUE,
neighbor_metric = "euclidean",
neighbor_k = 20L,
cluster_algorithm = "louvain",
cluster_resolution = 0.6,
seed = 11,
...
)Arguments
- srt_merge
A merged Seurat object that includes the batch information.
- batch
A character string specifying the batch variable name.
- append
Logical, if TRUE, the integrated data will be appended to the original Seurat object (srt_merge).
- srt_list
A list of Seurat objects to be checked and preprocessed.
- assay
The name of the assay to be used for downstream analysis.
- integration_method
A character string specifying the integration method to use. Supported methods are:
"Uncorrected","Seurat","scVI","MNN","fastMNN","Harmony","Scanorama","BBKNN","CSS","LIGER","Conos","ComBat". Default is"Uncorrected".- do_normalization
A logical value indicating whether data normalization should be performed.
- normalization_method
The normalization method to be used. Possible values are "LogNormalize", "SCT", and "TFIDF". Default is "LogNormalize".
- do_HVF_finding
A logical value indicating whether highly variable feature (HVF) finding should be performed. Default is TRUE.
- HVF_source
The source of highly variable features. Possible values are "global" and "separate". Default is "separate".
- HVF_method
The method for selecting highly variable features. Default is "vst".
- nHVF
The number of highly variable features to select. Default is 2000.
- HVF_min_intersection
The feature needs to be present in batches for a minimum number of times in order to be considered as highly variable. The default value is 1.
- HVF
A vector of highly variable features. Default is NULL.
- do_scaling
A logical value indicating whether to perform scaling. If TRUE, the function will force to scale the data using the ScaleData function.
- vars_to_regress
A vector of variable names to include as additional regression variables. Default is NULL.
- regression_model
The regression model to use for scaling. Options are "linear", "poisson", or "negativebinomial" (default is "linear").
- scale_within_batch
Whether to scale data within each batch. Only valid when the
integration_methodis one of"Uncorrected","Seurat","MNN","Harmony","BBKNN","CSS","ComBat".- linear_reduction
The linear dimensionality reduction method to use. Options are "pca", "svd", "ica", "nmf", "mds", or "glmpca" (default is "pca").
- linear_reduction_dims
The number of dimensions to keep after linear dimensionality reduction (default is 50).
- linear_reduction_dims_use
The dimensions to use for downstream analysis. If NULL, all dimensions will be used.
- linear_reduction_params
A list of parameters to pass to the linear dimensionality reduction method.
- force_linear_reduction
A logical value indicating whether to force linear dimensionality reduction even if the specified reduction is already present in the Seurat object.
- nonlinear_reduction
The nonlinear dimensionality reduction method to use. Options are "umap","umap-naive", "tsne", "dm", "phate", "pacmap", "trimap", "largevis", or "fr" (default is "umap").
- nonlinear_reduction_dims
The number of dimensions to keep after nonlinear dimensionality reduction. If a vector is provided, different numbers of dimensions can be specified for each method (default is c(2, 3)).
- nonlinear_reduction_params
A list of parameters to pass to the nonlinear dimensionality reduction method.
- force_nonlinear_reduction
A logical value indicating whether to force nonlinear dimensionality reduction even if the specified reduction is already present in the Seurat object.
- neighbor_metric
The distance metric to use for finding neighbors. Options are "euclidean", "cosine", "manhattan", or "hamming" (default is "euclidean").
- neighbor_k
The number of nearest neighbors to use for finding neighbors (default is 20).
- cluster_algorithm
The clustering algorithm to use. Options are "louvain", "slm", or "leiden" (default is "louvain").
- cluster_resolution
The resolution parameter to use for clustering. Larger values result in fewer clusters (default is 0.6).
- seed
An integer specifying the random seed for reproducibility. Default is 11.
- ...
Additional arguments to be passed to the integration method function.
Examples
data(panc8_sub)
panc8_sub <- integration_scop(
panc8_sub,
batch = "tech",
integration_method = "Uncorrected"
)
#> ℹ [2025-07-26 07:18:13] Start Uncorrected_integrate
#> ℹ [2025-07-26 07:18:13] Spliting srt_merge into srt_list by column tech...
#> ℹ [2025-07-26 07:18:13] Checking srt_list...
#> ℹ [2025-07-26 07:18:14] Data 1/5 of the srt_list has been log-normalized.
#> ℹ [2025-07-26 07:18:14] Perform FindVariableFeatures on the data 1/5 of the srt_list...
#> ℹ [2025-07-26 07:18:14] Data 2/5 of the srt_list has been log-normalized.
#> ℹ [2025-07-26 07:18:14] Perform FindVariableFeatures on the data 2/5 of the srt_list...
#> ℹ [2025-07-26 07:18:15] Data 3/5 of the srt_list has been log-normalized.
#> ℹ [2025-07-26 07:18:15] Perform FindVariableFeatures on the data 3/5 of the srt_list...
#> ℹ [2025-07-26 07:18:15] Data 4/5 of the srt_list has been log-normalized.
#> ℹ [2025-07-26 07:18:15] Perform FindVariableFeatures on the data 4/5 of the srt_list...
#> ℹ [2025-07-26 07:18:16] Data 5/5 of the srt_list has been log-normalized.
#> ℹ [2025-07-26 07:18:16] Perform FindVariableFeatures on the data 5/5 of the srt_list...
#> ℹ [2025-07-26 07:18:16] Use the separate HVF from srt_list...
#> ℹ [2025-07-26 07:18:16] Number of available HVF: 2000
#> ℹ [2025-07-26 07:18:17] Finished checking.
#> ℹ [2025-07-26 07:18:19] Perform integration(Uncorrected) on the data...
#> ℹ [2025-07-26 07:18:19] Perform ScaleData on the data...
#> ℹ [2025-07-26 07:18:19] Perform linear dimension reduction (pca) on the data...
#> ℹ [2025-07-26 07:18:19] Linear_reduction(pca) is already existed. Skip calculation.
#> Warning: The following arguments are not used: features
#> Warning: multiple layers are identified by counts.1.1.1.1 counts.2.1 counts.2.1.1 counts.2.1.1.1 data.1.1.1.1 scale.data.1.1.1.1 data.2.1.1.1 scale.data.2.1.1.1 data.2.1.1 scale.data.2.1.1 data.2.1 scale.data.2.1
#> only the first layer is used
#> ! [2025-07-26 07:18:19] Error: Not compatible with requested type: [type=S4; target=double].
#> ! [2025-07-26 07:18:19] Error when performing FindClusters. Skip this step...
#> ℹ [2025-07-26 07:18:19] Perform nonlinear dimension reduction (umap) on the data...
#> Warning: no non-missing arguments to min; returning Inf
#> Warning: no non-missing arguments to max; returning -Inf
#> ℹ [2025-07-26 07:18:19] Non-linear dimensionality reduction(umap) using Reduction(Uncorrectedpca, dims:Inf--Inf) as input
#> ! [2025-07-26 07:18:19] Error in `object[[graph]]`:
#> ! [2025-07-26 07:18:19] ! ‘Uncorrected_SNN’ not found in this Seurat object
#> ! [2025-07-26 07:18:19]
#> ! [2025-07-26 07:18:19] Error when performing nonlinear dimension reduction. Skip this step...
#> ℹ [2025-07-26 07:18:22] Uncorrected_integrate done
#> ℹ [2025-07-26 07:18:22] Elapsed time: 9.31 secs
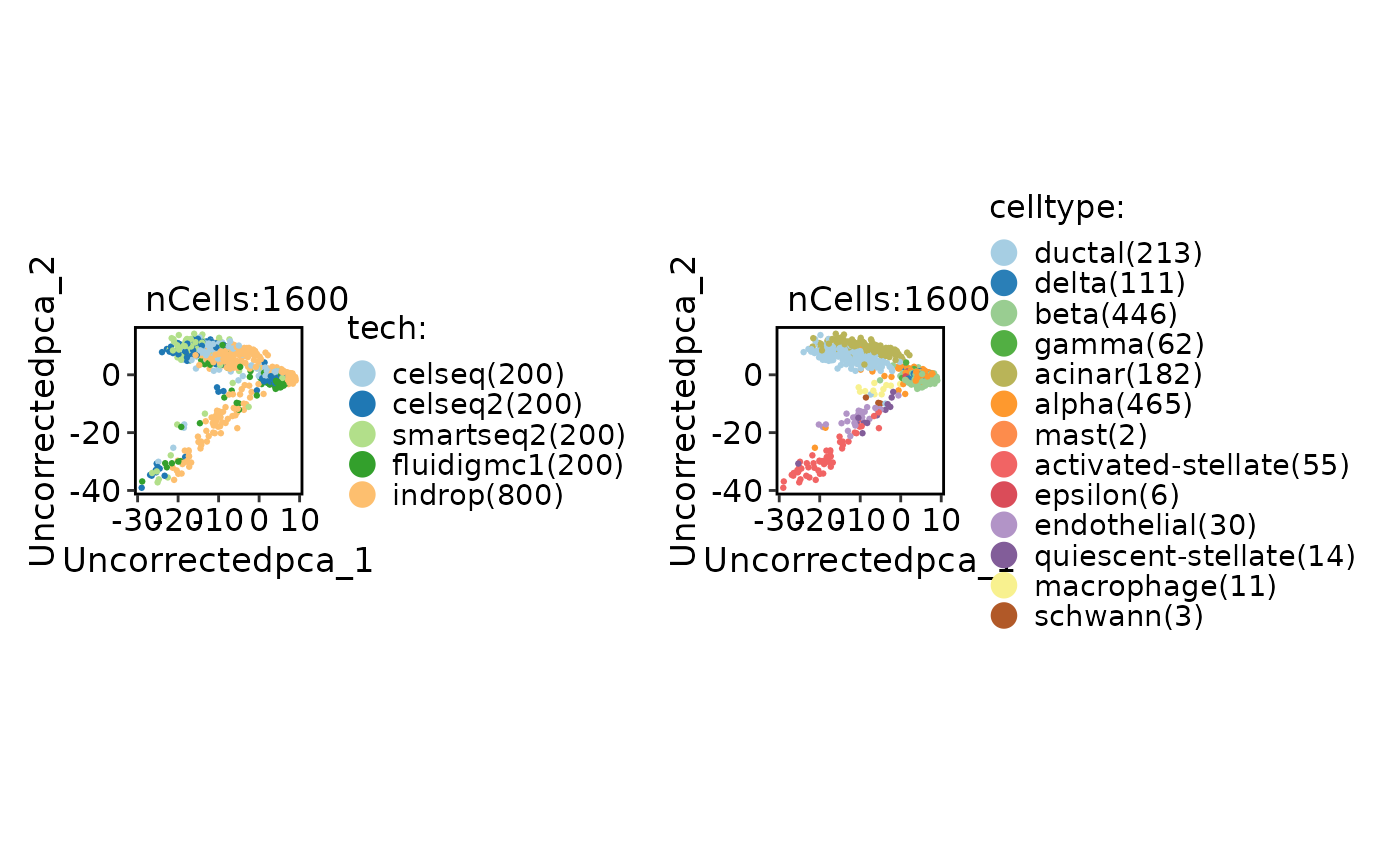
CellDimPlot(
panc8_sub,
group.by = c("tech", "celltype")
)
#> Warning: No shared levels found between `names(values)` of the manual scale and the
#> data's fill values.
#> Warning: No shared levels found between `names(values)` of the manual scale and the
#> data's fill values.
 panc8_sub <- integration_scop(
panc8_sub,
batch = "tech",
integration_method = "Uncorrected",
HVF_min_intersection = 5
)
#> ℹ [2025-07-26 07:18:22] Start Uncorrected_integrate
#> ℹ [2025-07-26 07:18:22] Spliting srt_merge into srt_list by column tech...
#> ℹ [2025-07-26 07:18:24] Checking srt_list...
#> Warning: The following features were labelled as variable in 'var.features' but had no corresponding rank in `var.features.rank` and will therefore be ignored: 'ABCC3', 'ABCC8', 'ABCC9', 'ABCG1', 'ABCG2', 'ABHD2', 'ABHD4', 'ABI3', 'ACHE', 'ACP5', 'ACSL4', 'ACSS1', 'ACTN1', 'ACTN4', 'ADAM8', 'ADAM9', 'ADAMTS5', 'ADAMTS9', 'ADAMTSL2', 'ADAP2', 'ADCY1', 'ADCY3', 'ADCY5', 'ADM2', 'ADRA2A', 'ADRBK2', 'AEN', 'AGR2', 'AGRN', 'AGTRAP', 'AHNAK', 'AHR', 'AKAP7', 'ALDH1A1', 'ALDH1A2', 'ALDH1B1', 'ALOX5AP', 'ALS2CL', 'AMOTL2', 'AMT', 'ANK1', 'ANKRD1', 'ANKRD37', 'ANLN', 'ANO6', 'ANXA13', 'ANXA3', 'ANXA9', 'AP5Z1', 'AQP3', 'ARC', 'ARG2', 'ARHGAP22', 'ARHGAP23', 'ARHGAP29', 'ARHGEF10L', 'ARHGEF2', 'ARHGEF39', 'ARHGEF40', 'ARL14', 'ARL4C', 'ARL6IP1', 'ARNTL2', 'ARX', 'ASAP1', 'ASAP2', 'ASCL2', 'ASF1B', 'ASGR1', 'ASNS', 'ASRGL1', 'ASS1', 'ATF3', 'ATF5', 'ATP11A', 'ATP1A1', 'ATP1B1', 'ATP1B2', 'ATP2A3', 'ATP8A1', 'B4GALT1', 'B4GALT5', 'BACE2', 'BAIAP2L1', 'BAIAP3', 'BAMBI', 'BBC3', 'BCL2L15', 'BCL3', 'BHLHE40', 'BHLHE41', 'BIK', 'BIRC5', 'BLNK', 'BMF', 'BMP1', 'BMP2', 'BMP7', 'BST2', 'BUB1B', 'BUB1', 'C1QA', 'C1QC', 'C1QL1', 'C2CD4A', 'C2CD4B', 'C2', 'CACNA1A', 'CACNA2D1', 'CACNG4', 'CADM2', 'CADPS', 'CALY', 'CAMK2B', 'CAPG', 'CAPN5', 'CAPN6', 'CARTPT', 'CASP9', 'CAV2', 'CBLB', 'CBLN4', 'CBS', 'CCBE1', 'CCDC34', 'CCDC51', 'CCDC69', 'CCNA1', 'CD14', 'CD276', 'CD300A', 'CD68', 'CD82', 'CD9', 'CDC20', 'CDC42EP3', 'CDCA8', 'CDH17', 'CDH3', 'CDK1', 'CDK5R2', 'CDKN1A', 'CDKN2A', 'CDKN2B', 'CDKN3', 'CDR2L', 'CDX2', 'CEBPB', 'CELF3', 'CENPW', 'CERCAM', 'CFI', 'CHAC1', 'CHDH', 'CHGB', 'CHRNA3', 'CITED2', 'CITED4', 'CKAP2', 'CKAP5', 'CKMT2', 'CKS1B', 'CKS2', 'CLCF1', 'CLDN1', 'CLIC4', 'CLMN', 'CLSPN', 'CMPK2', 'CMTM7', 'CNIH2', 'CNKSR2', 'CNN2', 'CNN3', 'CNTN1', 'COL13A1', 'COL16A1', 'COL18A1', 'COL4A2', 'COL5A3', 'COL9A2', 'COPG1', 'COPZ2', 'CORO1C', 'COTL1', 'COX7A1', 'CPLX2', 'CPPED1', 'CPVL', 'CRABP2', 'CRAT', 'CREB3L1', 'CREB5', 'CRIP1', 'CRLF1', 'CRTAC1', 'CRTAP', 'CRYM', 'CSF1R', 'CSGALNACT1', 'CTSC', 'CTSH', 'CTSO', 'CTSZ', 'CWC25', 'CX3CL1', 'CXADR', 'CXCL12', 'CXCL16', 'CXCL1', 'CYB5A', 'CYP26B1', 'CYP27A1', 'CYTH3', 'DAB2IP', 'DACH1', 'DACT1', 'DAPP1', 'DBN1', 'DBNDD1', 'DCUN1D2', 'DDC', 'DDIT4', 'DDX51', 'DEDD2', 'DHODH', 'DHRS2', 'DIP2A', 'DIRAS2', 'DNAJC12', 'DNAL1', 'DOCK4', 'DOCK6', 'DOCK8', 'DOCK9', 'DOK5', 'DPP4', 'DPP6', 'DSC2', 'DSEL', 'DUSP1', 'DUSP23', 'DUSP26', 'DUSP5', 'ECEL1', 'ECM1', 'ECT2', 'EDN2', 'EEF1D', 'EEF2K', 'EFEMP2', 'EFHD2', 'EFNA1', 'EGFR', 'EGR1', 'EGR4', 'EHD2', 'EHD4', 'EHF', 'ELAVL4', 'EMILIN1', 'EMP3', 'ENAH', 'ENO2', 'ENPP2', 'EPHB2', 'EPHB4', 'EPS8', 'ERBB3', 'ERO1L', 'ERRFI1', 'ETS2', 'EVA1A', 'EVA1B', 'F2RL2', 'F2RL3', 'F2R', 'F5', 'FA2H', 'FAM105A', 'FAM107B', 'FAM129B', 'FAM134B', 'FAM171A1', 'FAM227A', 'FAM43A', 'FAM46A', 'FAM46C', 'FAM84A', 'FBLIM1', 'FBP1', 'FBXL20', 'FBXO2', 'FBXO32', 'FBXO5', 'FBXO6', 'FCER1G', 'FCGRT', 'FERMT1', 'FEV', 'FFAR4', 'FGB', 'FGF13', 'FGFR2', 'FGFR3', 'FHL2', 'FHOD1', 'FJX1', 'FKBP10', 'FKBP11', 'FKBP5', 'FLNA', 'FMNL3', 'FNDC4', 'FNIP2', 'FOSB', 'FOSL2', 'FOXA3', 'FOXC1', 'FOXM1', 'FOXQ1', 'FRZB', 'FSCN1', 'FSTL3', 'FTH1', 'FURIN', 'FUT8', 'FXYD3', 'FXYD5', 'FXYD6', 'G0S2', 'G6PC', 'GAD2', 'GADD45A', 'GADD45B', 'GALE', 'GALM', 'GALNT2', 'GAL', 'GAS6', 'GAS7', 'GATA2', 'GATA4', 'GATSL3', 'GCGR', 'GCH1', 'GCK', 'GCLC', 'GCNT1', 'GCNT3', 'GC', 'GDA', 'GEM', 'GFOD2', 'GFPT2', 'GFRA1', 'GJA1', 'GJB1', 'GK', 'GLIPR1', 'GLRX', 'GLS', 'GMNN', 'GNPAT', 'GOLM1', 'GPC1', 'GPC6', 'GPD1', 'GPR160', 'GPR161', 'GPSM1', 'GPX3', 'GPX8', 'GRASP', 'GREB1', 'GSN', 'GSTM5', 'GSTP1', 'GTF3C3', 'GTPBP10', 'GTSE1', 'GULP1', 'H19', 'HABP2', 'HAND2', 'HDAC9', 'HEPACAM2', 'HEPH', 'HERPUD1', 'HES1', 'HEYL', 'HHEX', 'HIF1A', 'HIF3A', 'HILPDA', 'HIST1H1C', 'HIST1H2BK', 'HIST2H2BE', 'HK1', 'HK2', 'HKDC1', 'HMGB2', 'HMGB3', 'HMGCR', 'HMGCS1', 'HMMR', 'HN1', 'HNF1B', 'HOGA1', 'HOMER3', 'HOPX', 'HOXB2', 'HPCAL1', 'HPGD', 'HPN', 'HS3ST1', 'HSD11B2', 'HSD17B11', 'HSP90B1', 'HSPB1', 'HSPB8', 'HSPH1', 'ID1', 'ID4', 'IDH1', 'IDH2', 'IER2', 'IER3', 'IFIT1', 'IFITM2', 'IFNLR1', 'IGSF10', 'IGSF1', 'IGSF3', 'IL11', 'IL18', 'IL1R2', 'IMPA2', 'INPP5F', 'INSIG1', 'INSM1', 'IP6K1', 'IPCEF1', 'IRAK4', 'IRF1', 'IRF8', 'IRS2', 'IRX1', 'IRX2', 'IRX4', 'ISL1', 'ITGA11', 'ITGA3', 'ITGA9', 'ITGAV', 'ITGB4', 'ITGB6', 'ITGB8', 'ITIH5', 'ITPRIPL2', 'JAG1', 'JARID2', 'JRK', 'JSRP1', 'JUN', 'KANK1', 'KANK2', 'KCNA5', 'KCNAB1', 'KCNE3', 'KCNG1', 'KCNH6', 'KCNJ11', 'KCNJ16', 'KCNJ5', 'KCNJ6', 'KCNK16', 'KCNK3', 'KCNK5', 'KCNMA1', 'KCNN3', 'KCNQ1OT1', 'KCNQ1', 'KCTD12', 'KCTD8', 'KDELC2', 'KDELR3', 'KDM5D', 'KIF11', 'KIF12', 'KIF14', 'KIF21B', 'KIF2C', 'KIF5C', 'KISS1R', 'KLF2', 'KLF4', 'KLF5', 'KLF6', 'KLHDC8A', 'KLHL21', 'KLK11', 'KNSTRN', 'KPNA2', 'KRT17', 'KRTCAP3', 'L2HGDH', 'LAD1', 'LAMB1', 'LAMB2', 'LAPTM5', 'LBH', 'LDLR', 'LECT1', 'LEFTY1', 'LGALS3', 'LGALS9', 'LGI2', 'LHFP', 'LIMA1', 'LIMCH1', 'LIMK2', 'LIMS1', 'LIPH', 'LMO7', 'LONRF2', 'LOXL2', 'LPAR6', 'LRP12', 'LRP1', 'LRRC10B', 'LRRC59', 'LSAMP', 'LTBP4', 'LURAP1L', 'LUZP1', 'LXN', 'LY6E', 'LY6H', 'LYN', 'LYPD1', 'LYPD6B', 'MACC1', 'MAFA', 'MAFB', 'MAFF', 'MAL2', 'MALL', 'MAN1C1', 'MANF', 'MAOB', 'MAP1B', 'MAP2', 'MAP7D2', 'MAPT', 'MARCKSL1', 'MATN2', 'MDFI', 'MDK', 'MECOM', 'MEIS2', 'MERTK', 'METRN', 'METTL20', 'METTL22', 'MFI2', 'MGAT4A', 'MGAT4B', 'MGLL', 'MICAL2', 'MID1IP1', 'MIR143HG', 'MLLT11', 'MLXIPL', 'MMP14', 'MNX1', 'MPZL2', 'MRC1', 'MRC2', 'MSMO1', 'MSN', 'MSRB1', 'MSRB3', 'MTUS1', 'MUM1L1', 'MYC', 'MYH10', 'MYH9', 'MYL12A', 'MYO10', 'MYO1E', 'MYO5B', 'MYRF', 'MYZAP', 'NAA16', 'NCAM1', 'NCAPG', 'NCAPH', 'NCMAP', 'NDRG1', 'NDRG2', 'NDRG4', 'NDUFA4L2', 'NDUFA4', 'NEK8', 'NEURL1B', 'NEURL3', 'NEUROD1', 'NFATC4', 'NFKBIA', 'NFKBIE', 'NINJ1', 'NKD1', 'NKX2-2', 'NLN', 'NMB', 'NMNAT1', 'NOB1', 'NOTCH1', 'NOTCH2', 'NOTCH3', 'NOTCH4', 'NOX4', 'NPHS1', 'NPNT', 'NPTX2', 'NQO1', 'NR0B1', 'NR0B2', 'NR1D1', 'NR2F2', 'NR4A2', 'NR5A2', 'NRP2', 'NTN4', 'NUAK2', 'NUF2', 'NUSAP1', 'OCLN', 'OGDHL', 'OLFML2B', 'OPA3', 'ORM1', 'P4HA3', 'PACSIN2', 'PAH', 'PAK3', 'PALLD', 'PAMR1', 'PAPSS2', 'PARM1', 'PAX6', 'PBK', 'PCDH17', 'PCDH18', 'PCDH9', 'PCK1', 'PCNA', 'PCP4', 'PCSK2', 'PCSK5', 'PDE3A', 'PDE3B', 'PDE8B', 'PDGFA', 'PDGFD', 'PDIA4', 'PDLIM1', 'PDLIM4', 'PDLIM7', 'PDP2', 'PDX1', 'PEMT', 'PER3', 'PEX13', 'PFKFB3', 'PGBD5', 'PGM1', 'PGM2L1', 'PHF19', 'PHGDH', 'PHGR1', 'PHLDA2', 'PHLDB1', 'PID1', 'PIEZO1', 'PIK3AP1', 'PIK3R1', 'PIM1', 'PKIB', 'PKNOX2', 'PKP2', 'PLA2G4C', 'PLA2G7', 'PLAA', 'PLAGL1', 'PLAT', 'PLCXD3', 'PLEKHA2', 'PLEKHB1', 'PLEKHG2', 'PLEKHO1', 'PLIN5', 'PLK2', 'PLLP', 'PLOD2', 'PLSCR1', 'PLXDC1', 'PLXNA1', 'PLXNA2', 'PLXND1', 'PMAIP1', 'PNMA2', 'PNRC1', 'POC1A', 'POLD4', 'PON2', 'POU2F2', 'PPARGC1A', 'PPIC', 'PPIF', 'PPL', 'PPP1R15A', 'PPP1R18', 'PPP1R1A', 'PPP1R1B', 'PPP2R2B', 'PRC1', 'PRELP', 'PRKCDBP', 'PRKG1', 'PRKX', 'PRLHR', 'PRMT6', 'PRR11', 'PRR15L', 'PRSS22', 'PRSS23', 'PRSS8', 'PTAFR', 'PTGDS', 'PTGES', 'PTGFR', 'PTK7', 'PTPN14', 'PTPN3', 'PTPRN', 'PTPRT', 'PTRF', 'PTTG1', 'PVRL3', 'PVRL4', 'PYCARD', 'PYCR1', 'PYROXD2', 'QPRT', 'QSOX1', 'RAB11FIP1', 'RAB33A', 'RAB3B', 'RAB42', 'RACGAP1', 'RAMP1', 'RAP1GAP', 'RAPGEF5', 'RARG', 'RARRES1', 'RASA3', 'RASEF', 'RASGRP3', 'RASL12', 'RASSF4', 'RASSF9', 'RBM38', 'RBP1', 'RBPMS', 'RCL1', 'RCN3', 'RDH10', 'RDX', 'RELB', 'RETSAT', 'RFTN1', 'RFX6', 'RGS5', 'RHBDF1', 'RHOB', 'RHOC', 'RHOD', 'RHOV', 'RHPN2', 'RINL', 'RNASE6', 'RNF114', 'RNF157', 'RNF24', 'RNF32', 'ROBO1', 'ROR2', 'RPL22L1', 'RPS6KA2', 'RRAGD', 'RSU1', 'S100A10', 'S100A11', 'S100A16', 'S1PR3', 'SAMHD1', 'SCG2', 'SCG3', 'SCN1B', 'SCNN1A', 'SCTR', 'SDC2', 'SDS', 'SEL1L3', 'SEMA3B', 'SEMA3C', 'SEMA4B', 'SEMA6A', 'SEMA7A', 'SEPT3', 'SEPT6', 'SEPT9', 'SERPINA6', 'SERPINE2', 'SERPINF1', 'SERPINI1', 'SERTM1', 'SETX', 'SEZ6L', 'SFMBT2', 'SFN', 'SFRP1', 'SFRP5', 'SGOL1', 'SGSM1', 'SGSM3', 'SH3BGRL3', 'SH3BP4', 'SH3KBP1', 'SH3PXD2A', 'SHANK2', 'SHC1', 'SHROOM1', 'SHROOM3', 'SIRPA', 'SIX4', 'SKIL', 'SLC12A2', 'SLC12A7', 'SLC15A4', 'SLC16A3', 'SLC16A5', 'SLC16A7', 'SLC16A9', 'SLC1A5', 'SLC22A17', 'SLC22A3', 'SLC25A43', 'SLC26A4', 'SLC29A4', 'SLC2A13', 'SLC30A8', 'SLC38A1', 'SLC38A2', 'SLC38A5', 'SLC39A14', 'SLC39A5', 'SLC40A1', 'SLC43A3', 'SLC44A2', 'SLC45A3', 'SLC4A4', 'SLC4A5', 'SL
#> ℹ [2025-07-26 07:18:24] Data 1/5 of the srt_list has been log-normalized.
#> ℹ [2025-07-26 07:18:24] Perform FindVariableFeatures on the data 1/5 of the srt_list...
#> Warning: The following features were labelled as variable in 'var.features' but had no corresponding rank in `var.features.rank` and will therefore be ignored: 'ABCC3', 'ABCC8', 'ABCC9', 'ABCG1', 'ABCG2', 'ABHD2', 'ABHD4', 'ABI3', 'ACHE', 'ACP5', 'ACSL4', 'ACSS1', 'ACTN1', 'ACTN4', 'ADAM8', 'ADAM9', 'ADAMTS5', 'ADAMTS9', 'ADAMTSL2', 'ADAP2', 'ADCY1', 'ADCY3', 'ADCY5', 'ADM2', 'ADRA2A', 'ADRBK2', 'AEN', 'AGR2', 'AGRN', 'AGTRAP', 'AHNAK', 'AHR', 'AKAP7', 'ALDH1A1', 'ALDH1A2', 'ALDH1B1', 'ALOX5AP', 'ALS2CL', 'AMOTL2', 'AMT', 'ANK1', 'ANKRD1', 'ANKRD37', 'ANLN', 'ANO6', 'ANXA13', 'ANXA3', 'ANXA9', 'AP5Z1', 'AQP3', 'ARC', 'ARG2', 'ARHGAP22', 'ARHGAP23', 'ARHGAP29', 'ARHGEF10L', 'ARHGEF2', 'ARHGEF39', 'ARHGEF40', 'ARL14', 'ARL4C', 'ARL6IP1', 'ARNTL2', 'ARX', 'ASAP1', 'ASAP2', 'ASCL2', 'ASF1B', 'ASGR1', 'ASNS', 'ASRGL1', 'ASS1', 'ATF3', 'ATF5', 'ATP11A', 'ATP1A1', 'ATP1B1', 'ATP1B2', 'ATP2A3', 'ATP8A1', 'B4GALT1', 'B4GALT5', 'BACE2', 'BAIAP2L1', 'BAIAP3', 'BAMBI', 'BBC3', 'BCL2L15', 'BCL3', 'BHLHE40', 'BHLHE41', 'BIK', 'BIRC5', 'BLNK', 'BMF', 'BMP1', 'BMP2', 'BMP7', 'BST2', 'BUB1B', 'BUB1', 'C1QA', 'C1QC', 'C1QL1', 'C2CD4A', 'C2CD4B', 'C2', 'CACNA1A', 'CACNA2D1', 'CACNG4', 'CADM2', 'CADPS', 'CALY', 'CAMK2B', 'CAPG', 'CAPN5', 'CAPN6', 'CARTPT', 'CASP9', 'CAV2', 'CBLB', 'CBLN4', 'CBS', 'CCBE1', 'CCDC34', 'CCDC51', 'CCDC69', 'CCNA1', 'CD14', 'CD276', 'CD300A', 'CD68', 'CD82', 'CD9', 'CDC20', 'CDC42EP3', 'CDCA8', 'CDH17', 'CDH3', 'CDK1', 'CDK5R2', 'CDKN1A', 'CDKN2A', 'CDKN2B', 'CDKN3', 'CDR2L', 'CDX2', 'CEBPB', 'CELF3', 'CENPW', 'CERCAM', 'CFI', 'CHAC1', 'CHDH', 'CHGB', 'CHRNA3', 'CITED2', 'CITED4', 'CKAP2', 'CKAP5', 'CKMT2', 'CKS1B', 'CKS2', 'CLCF1', 'CLDN1', 'CLIC4', 'CLMN', 'CLSPN', 'CMPK2', 'CMTM7', 'CNIH2', 'CNKSR2', 'CNN2', 'CNN3', 'CNTN1', 'COL13A1', 'COL16A1', 'COL18A1', 'COL4A2', 'COL5A3', 'COL9A2', 'COPG1', 'COPZ2', 'CORO1C', 'COTL1', 'COX7A1', 'CPLX2', 'CPPED1', 'CPVL', 'CRABP2', 'CRAT', 'CREB3L1', 'CREB5', 'CRIP1', 'CRLF1', 'CRTAC1', 'CRTAP', 'CRYM', 'CSF1R', 'CSGALNACT1', 'CTSC', 'CTSH', 'CTSO', 'CTSZ', 'CWC25', 'CX3CL1', 'CXADR', 'CXCL12', 'CXCL16', 'CXCL1', 'CYB5A', 'CYP26B1', 'CYP27A1', 'CYTH3', 'DAB2IP', 'DACH1', 'DACT1', 'DAPP1', 'DBN1', 'DBNDD1', 'DCUN1D2', 'DDC', 'DDIT4', 'DDX51', 'DEDD2', 'DHODH', 'DHRS2', 'DIP2A', 'DIRAS2', 'DNAJC12', 'DNAL1', 'DOCK4', 'DOCK6', 'DOCK8', 'DOCK9', 'DOK5', 'DPP4', 'DPP6', 'DSC2', 'DSEL', 'DUSP1', 'DUSP23', 'DUSP26', 'DUSP5', 'ECEL1', 'ECM1', 'ECT2', 'EDN2', 'EEF1D', 'EEF2K', 'EFEMP2', 'EFHD2', 'EFNA1', 'EGFR', 'EGR1', 'EGR4', 'EHD2', 'EHD4', 'EHF', 'ELAVL4', 'EMILIN1', 'EMP3', 'ENAH', 'ENO2', 'ENPP2', 'EPHB2', 'EPHB4', 'EPS8', 'ERBB3', 'ERO1L', 'ERRFI1', 'ETS2', 'EVA1A', 'EVA1B', 'F2RL2', 'F2RL3', 'F2R', 'F5', 'FA2H', 'FAM105A', 'FAM107B', 'FAM129B', 'FAM134B', 'FAM171A1', 'FAM227A', 'FAM43A', 'FAM46A', 'FAM46C', 'FAM84A', 'FBLIM1', 'FBP1', 'FBXL20', 'FBXO2', 'FBXO32', 'FBXO5', 'FBXO6', 'FCER1G', 'FCGRT', 'FERMT1', 'FEV', 'FFAR4', 'FGB', 'FGF13', 'FGFR2', 'FGFR3', 'FHL2', 'FHOD1', 'FJX1', 'FKBP10', 'FKBP11', 'FKBP5', 'FLNA', 'FMNL3', 'FNDC4', 'FNIP2', 'FOSB', 'FOSL2', 'FOXA3', 'FOXC1', 'FOXM1', 'FOXQ1', 'FRZB', 'FSCN1', 'FSTL3', 'FTH1', 'FURIN', 'FUT8', 'FXYD3', 'FXYD5', 'FXYD6', 'G0S2', 'G6PC', 'GAD2', 'GADD45A', 'GADD45B', 'GALE', 'GALM', 'GALNT2', 'GAL', 'GAS6', 'GAS7', 'GATA2', 'GATA4', 'GATSL3', 'GCGR', 'GCH1', 'GCK', 'GCLC', 'GCNT1', 'GCNT3', 'GC', 'GDA', 'GEM', 'GFOD2', 'GFPT2', 'GFRA1', 'GJA1', 'GJB1', 'GK', 'GLIPR1', 'GLRX', 'GLS', 'GMNN', 'GNPAT', 'GOLM1', 'GPC1', 'GPC6', 'GPD1', 'GPR160', 'GPR161', 'GPSM1', 'GPX3', 'GPX8', 'GRASP', 'GREB1', 'GSN', 'GSTM5', 'GSTP1', 'GTF3C3', 'GTPBP10', 'GTSE1', 'GULP1', 'H19', 'HABP2', 'HAND2', 'HDAC9', 'HEPACAM2', 'HEPH', 'HERPUD1', 'HES1', 'HEYL', 'HHEX', 'HIF1A', 'HIF3A', 'HILPDA', 'HIST1H1C', 'HIST1H2BK', 'HIST2H2BE', 'HK1', 'HK2', 'HKDC1', 'HMGB2', 'HMGB3', 'HMGCR', 'HMGCS1', 'HMMR', 'HN1', 'HNF1B', 'HOGA1', 'HOMER3', 'HOPX', 'HOXB2', 'HPCAL1', 'HPGD', 'HPN', 'HS3ST1', 'HSD11B2', 'HSD17B11', 'HSP90B1', 'HSPB1', 'HSPB8', 'HSPH1', 'ID1', 'ID4', 'IDH1', 'IDH2', 'IER2', 'IER3', 'IFIT1', 'IFITM2', 'IFNLR1', 'IGSF10', 'IGSF1', 'IGSF3', 'IL11', 'IL18', 'IL1R2', 'IMPA2', 'INPP5F', 'INSIG1', 'INSM1', 'IP6K1', 'IPCEF1', 'IRAK4', 'IRF1', 'IRF8', 'IRS2', 'IRX1', 'IRX2', 'IRX4', 'ISL1', 'ITGA11', 'ITGA3', 'ITGA9', 'ITGAV', 'ITGB4', 'ITGB6', 'ITGB8', 'ITIH5', 'ITPRIPL2', 'JAG1', 'JARID2', 'JRK', 'JSRP1', 'JUN', 'KANK1', 'KANK2', 'KCNA5', 'KCNAB1', 'KCNE3', 'KCNG1', 'KCNH6', 'KCNJ11', 'KCNJ16', 'KCNJ5', 'KCNJ6', 'KCNK16', 'KCNK3', 'KCNK5', 'KCNMA1', 'KCNN3', 'KCNQ1OT1', 'KCNQ1', 'KCTD12', 'KCTD8', 'KDELC2', 'KDELR3', 'KDM5D', 'KIF11', 'KIF12', 'KIF14', 'KIF21B', 'KIF2C', 'KIF5C', 'KISS1R', 'KLF2', 'KLF4', 'KLF5', 'KLF6', 'KLHDC8A', 'KLHL21', 'KLK11', 'KNSTRN', 'KPNA2', 'KRT17', 'KRTCAP3', 'L2HGDH', 'LAD1', 'LAMB1', 'LAMB2', 'LAPTM5', 'LBH', 'LDLR', 'LECT1', 'LEFTY1', 'LGALS3', 'LGALS9', 'LGI2', 'LHFP', 'LIMA1', 'LIMCH1', 'LIMK2', 'LIMS1', 'LIPH', 'LMO7', 'LONRF2', 'LOXL2', 'LPAR6', 'LRP12', 'LRP1', 'LRRC10B', 'LRRC59', 'LSAMP', 'LTBP4', 'LURAP1L', 'LUZP1', 'LXN', 'LY6E', 'LY6H', 'LYN', 'LYPD1', 'LYPD6B', 'MACC1', 'MAFA', 'MAFB', 'MAFF', 'MAL2', 'MALL', 'MAN1C1', 'MANF', 'MAOB', 'MAP1B', 'MAP2', 'MAP7D2', 'MAPT', 'MARCKSL1', 'MATN2', 'MDFI', 'MDK', 'MECOM', 'MEIS2', 'MERTK', 'METRN', 'METTL20', 'METTL22', 'MFI2', 'MGAT4A', 'MGAT4B', 'MGLL', 'MICAL2', 'MID1IP1', 'MIR143HG', 'MLLT11', 'MLXIPL', 'MMP14', 'MNX1', 'MPZL2', 'MRC1', 'MRC2', 'MSMO1', 'MSN', 'MSRB1', 'MSRB3', 'MTUS1', 'MUM1L1', 'MYC', 'MYH10', 'MYH9', 'MYL12A', 'MYO10', 'MYO1E', 'MYO5B', 'MYRF', 'MYZAP', 'NAA16', 'NCAM1', 'NCAPG', 'NCAPH', 'NCMAP', 'NDRG1', 'NDRG2', 'NDRG4', 'NDUFA4L2', 'NDUFA4', 'NEK8', 'NEURL1B', 'NEURL3', 'NEUROD1', 'NFATC4', 'NFKBIA', 'NFKBIE', 'NINJ1', 'NKD1', 'NKX2-2', 'NLN', 'NMB', 'NMNAT1', 'NOB1', 'NOTCH1', 'NOTCH2', 'NOTCH3', 'NOTCH4', 'NOX4', 'NPHS1', 'NPNT', 'NPTX2', 'NQO1', 'NR0B1', 'NR0B2', 'NR1D1', 'NR2F2', 'NR4A2', 'NR5A2', 'NRP2', 'NTN4', 'NUAK2', 'NUF2', 'NUSAP1', 'OCLN', 'OGDHL', 'OLFML2B', 'OPA3', 'ORM1', 'P4HA3', 'PACSIN2', 'PAH', 'PAK3', 'PALLD', 'PAMR1', 'PAPSS2', 'PARM1', 'PAX6', 'PBK', 'PCDH17', 'PCDH18', 'PCDH9', 'PCK1', 'PCNA', 'PCP4', 'PCSK2', 'PCSK5', 'PDE3A', 'PDE3B', 'PDE8B', 'PDGFA', 'PDGFD', 'PDIA4', 'PDLIM1', 'PDLIM4', 'PDLIM7', 'PDP2', 'PDX1', 'PEMT', 'PER3', 'PEX13', 'PFKFB3', 'PGBD5', 'PGM1', 'PGM2L1', 'PHF19', 'PHGDH', 'PHGR1', 'PHLDA2', 'PHLDB1', 'PID1', 'PIEZO1', 'PIK3AP1', 'PIK3R1', 'PIM1', 'PKIB', 'PKNOX2', 'PKP2', 'PLA2G4C', 'PLA2G7', 'PLAA', 'PLAGL1', 'PLAT', 'PLCXD3', 'PLEKHA2', 'PLEKHB1', 'PLEKHG2', 'PLEKHO1', 'PLIN5', 'PLK2', 'PLLP', 'PLOD2', 'PLSCR1', 'PLXDC1', 'PLXNA1', 'PLXNA2', 'PLXND1', 'PMAIP1', 'PNMA2', 'PNRC1', 'POC1A', 'POLD4', 'PON2', 'POU2F2', 'PPARGC1A', 'PPIC', 'PPIF', 'PPL', 'PPP1R15A', 'PPP1R18', 'PPP1R1A', 'PPP1R1B', 'PPP2R2B', 'PRC1', 'PRELP', 'PRKCDBP', 'PRKG1', 'PRKX', 'PRLHR', 'PRMT6', 'PRR11', 'PRR15L', 'PRSS22', 'PRSS23', 'PRSS8', 'PTAFR', 'PTGDS', 'PTGES', 'PTGFR', 'PTK7', 'PTPN14', 'PTPN3', 'PTPRN', 'PTPRT', 'PTRF', 'PTTG1', 'PVRL3', 'PVRL4', 'PYCARD', 'PYCR1', 'PYROXD2', 'QPRT', 'QSOX1', 'RAB11FIP1', 'RAB33A', 'RAB3B', 'RAB42', 'RACGAP1', 'RAMP1', 'RAP1GAP', 'RAPGEF5', 'RARG', 'RARRES1', 'RASA3', 'RASEF', 'RASGRP3', 'RASL12', 'RASSF4', 'RASSF9', 'RBM38', 'RBP1', 'RBPMS', 'RCL1', 'RCN3', 'RDH10', 'RDX', 'RELB', 'RETSAT', 'RFTN1', 'RFX6', 'RGS5', 'RHBDF1', 'RHOB', 'RHOC', 'RHOD', 'RHOV', 'RHPN2', 'RINL', 'RNASE6', 'RNF114', 'RNF157', 'RNF24', 'RNF32', 'ROBO1', 'ROR2', 'RPL22L1', 'RPS6KA2', 'RRAGD', 'RSU1', 'S100A10', 'S100A11', 'S100A16', 'S1PR3', 'SAMHD1', 'SCG2', 'SCG3', 'SCN1B', 'SCNN1A', 'SCTR', 'SDC2', 'SDS', 'SEL1L3', 'SEMA3B', 'SEMA3C', 'SEMA4B', 'SEMA6A', 'SEMA7A', 'SEPT3', 'SEPT6', 'SEPT9', 'SERPINA6', 'SERPINE2', 'SERPINF1', 'SERPINI1', 'SERTM1', 'SETX', 'SEZ6L', 'SFMBT2', 'SFN', 'SFRP1', 'SFRP5', 'SGOL1', 'SGSM1', 'SGSM3', 'SH3BGRL3', 'SH3BP4', 'SH3KBP1', 'SH3PXD2A', 'SHANK2', 'SHC1', 'SHROOM1', 'SHROOM3', 'SIRPA', 'SIX4', 'SKIL', 'SLC12A2', 'SLC12A7', 'SLC15A4', 'SLC16A3', 'SLC16A5', 'SLC16A7', 'SLC16A9', 'SLC1A5', 'SLC22A17', 'SLC22A3', 'SLC25A43', 'SLC26A4', 'SLC29A4', 'SLC2A13', 'SLC30A8', 'SLC38A1', 'SLC38A2', 'SLC38A5', 'SLC39A14', 'SLC39A5', 'SLC40A1', 'SLC43A3', 'SLC44A2', 'SLC45A3', 'SLC4A4', 'SLC4A5', 'SL
#> ℹ [2025-07-26 07:18:25] Data 2/5 of the srt_list has been log-normalized.
#> ℹ [2025-07-26 07:18:25] Perform FindVariableFeatures on the data 2/5 of the srt_list...
#> Warning: The following features were labelled as variable in 'var.features' but had no corresponding rank in `var.features.rank` and will therefore be ignored: 'ABCC3', 'ABCC8', 'ABCC9', 'ABCG1', 'ABCG2', 'ABHD2', 'ABHD4', 'ABI3', 'ACHE', 'ACP5', 'ACSL4', 'ACSS1', 'ACTN1', 'ACTN4', 'ADAM8', 'ADAM9', 'ADAMTS5', 'ADAMTS9', 'ADAMTSL2', 'ADAP2', 'ADCY1', 'ADCY3', 'ADCY5', 'ADM2', 'ADRA2A', 'ADRBK2', 'AEN', 'AGR2', 'AGRN', 'AGTRAP', 'AHNAK', 'AHR', 'AKAP7', 'ALDH1A1', 'ALDH1A2', 'ALDH1B1', 'ALOX5AP', 'ALS2CL', 'AMOTL2', 'AMT', 'ANK1', 'ANKRD1', 'ANKRD37', 'ANLN', 'ANO6', 'ANXA13', 'ANXA3', 'ANXA9', 'AP5Z1', 'AQP3', 'ARC', 'ARG2', 'ARHGAP22', 'ARHGAP23', 'ARHGAP29', 'ARHGEF10L', 'ARHGEF2', 'ARHGEF39', 'ARHGEF40', 'ARL14', 'ARL4C', 'ARL6IP1', 'ARNTL2', 'ARX', 'ASAP1', 'ASAP2', 'ASCL2', 'ASF1B', 'ASGR1', 'ASNS', 'ASRGL1', 'ASS1', 'ATF3', 'ATF5', 'ATP11A', 'ATP1A1', 'ATP1B1', 'ATP1B2', 'ATP2A3', 'ATP8A1', 'B4GALT1', 'B4GALT5', 'BACE2', 'BAIAP2L1', 'BAIAP3', 'BAMBI', 'BBC3', 'BCL2L15', 'BCL3', 'BHLHE40', 'BHLHE41', 'BIK', 'BIRC5', 'BLNK', 'BMF', 'BMP1', 'BMP2', 'BMP7', 'BST2', 'BUB1B', 'BUB1', 'C1QA', 'C1QC', 'C1QL1', 'C2CD4A', 'C2CD4B', 'C2', 'CACNA1A', 'CACNA2D1', 'CACNG4', 'CADM2', 'CADPS', 'CALY', 'CAMK2B', 'CAPG', 'CAPN5', 'CAPN6', 'CARTPT', 'CASP9', 'CAV2', 'CBLB', 'CBLN4', 'CBS', 'CCBE1', 'CCDC34', 'CCDC51', 'CCDC69', 'CCNA1', 'CD14', 'CD276', 'CD300A', 'CD68', 'CD82', 'CD9', 'CDC20', 'CDC42EP3', 'CDCA8', 'CDH17', 'CDH3', 'CDK1', 'CDK5R2', 'CDKN1A', 'CDKN2A', 'CDKN2B', 'CDKN3', 'CDR2L', 'CDX2', 'CEBPB', 'CELF3', 'CENPW', 'CERCAM', 'CFI', 'CHAC1', 'CHDH', 'CHGB', 'CHRNA3', 'CITED2', 'CITED4', 'CKAP2', 'CKAP5', 'CKMT2', 'CKS1B', 'CKS2', 'CLCF1', 'CLDN1', 'CLIC4', 'CLMN', 'CLSPN', 'CMPK2', 'CMTM7', 'CNIH2', 'CNKSR2', 'CNN2', 'CNN3', 'CNTN1', 'COL13A1', 'COL16A1', 'COL18A1', 'COL4A2', 'COL5A3', 'COL9A2', 'COPG1', 'COPZ2', 'CORO1C', 'COTL1', 'COX7A1', 'CPLX2', 'CPPED1', 'CPVL', 'CRABP2', 'CRAT', 'CREB3L1', 'CREB5', 'CRIP1', 'CRLF1', 'CRTAC1', 'CRTAP', 'CRYM', 'CSF1R', 'CSGALNACT1', 'CTSC', 'CTSH', 'CTSO', 'CTSZ', 'CWC25', 'CX3CL1', 'CXADR', 'CXCL12', 'CXCL16', 'CXCL1', 'CYB5A', 'CYP26B1', 'CYP27A1', 'CYTH3', 'DAB2IP', 'DACH1', 'DACT1', 'DAPP1', 'DBN1', 'DBNDD1', 'DCUN1D2', 'DDC', 'DDIT4', 'DDX51', 'DEDD2', 'DHODH', 'DHRS2', 'DIP2A', 'DIRAS2', 'DNAJC12', 'DNAL1', 'DOCK4', 'DOCK6', 'DOCK8', 'DOCK9', 'DOK5', 'DPP4', 'DPP6', 'DSC2', 'DSEL', 'DUSP1', 'DUSP23', 'DUSP26', 'DUSP5', 'ECEL1', 'ECM1', 'ECT2', 'EDN2', 'EEF1D', 'EEF2K', 'EFEMP2', 'EFHD2', 'EFNA1', 'EGFR', 'EGR1', 'EGR4', 'EHD2', 'EHD4', 'EHF', 'ELAVL4', 'EMILIN1', 'EMP3', 'ENAH', 'ENO2', 'ENPP2', 'EPHB2', 'EPHB4', 'EPS8', 'ERBB3', 'ERO1L', 'ERRFI1', 'ETS2', 'EVA1A', 'EVA1B', 'F2RL2', 'F2RL3', 'F2R', 'F5', 'FA2H', 'FAM105A', 'FAM107B', 'FAM129B', 'FAM134B', 'FAM171A1', 'FAM227A', 'FAM43A', 'FAM46A', 'FAM46C', 'FAM84A', 'FBLIM1', 'FBP1', 'FBXL20', 'FBXO2', 'FBXO32', 'FBXO5', 'FBXO6', 'FCER1G', 'FCGRT', 'FERMT1', 'FEV', 'FFAR4', 'FGB', 'FGF13', 'FGFR2', 'FGFR3', 'FHL2', 'FHOD1', 'FJX1', 'FKBP10', 'FKBP11', 'FKBP5', 'FLNA', 'FMNL3', 'FNDC4', 'FNIP2', 'FOSB', 'FOSL2', 'FOXA3', 'FOXC1', 'FOXM1', 'FOXQ1', 'FRZB', 'FSCN1', 'FSTL3', 'FTH1', 'FURIN', 'FUT8', 'FXYD3', 'FXYD5', 'FXYD6', 'G0S2', 'G6PC', 'GAD2', 'GADD45A', 'GADD45B', 'GALE', 'GALM', 'GALNT2', 'GAL', 'GAS6', 'GAS7', 'GATA2', 'GATA4', 'GATSL3', 'GCGR', 'GCH1', 'GCK', 'GCLC', 'GCNT1', 'GCNT3', 'GC', 'GDA', 'GEM', 'GFOD2', 'GFPT2', 'GFRA1', 'GJA1', 'GJB1', 'GK', 'GLIPR1', 'GLRX', 'GLS', 'GMNN', 'GNPAT', 'GOLM1', 'GPC1', 'GPC6', 'GPD1', 'GPR160', 'GPR161', 'GPSM1', 'GPX3', 'GPX8', 'GRASP', 'GREB1', 'GSN', 'GSTM5', 'GSTP1', 'GTF3C3', 'GTPBP10', 'GTSE1', 'GULP1', 'H19', 'HABP2', 'HAND2', 'HDAC9', 'HEPACAM2', 'HEPH', 'HERPUD1', 'HES1', 'HEYL', 'HHEX', 'HIF1A', 'HIF3A', 'HILPDA', 'HIST1H1C', 'HIST1H2BK', 'HIST2H2BE', 'HK1', 'HK2', 'HKDC1', 'HMGB2', 'HMGB3', 'HMGCR', 'HMGCS1', 'HMMR', 'HN1', 'HNF1B', 'HOGA1', 'HOMER3', 'HOPX', 'HOXB2', 'HPCAL1', 'HPGD', 'HPN', 'HS3ST1', 'HSD11B2', 'HSD17B11', 'HSP90B1', 'HSPB1', 'HSPB8', 'HSPH1', 'ID1', 'ID4', 'IDH1', 'IDH2', 'IER2', 'IER3', 'IFIT1', 'IFITM2', 'IFNLR1', 'IGSF10', 'IGSF1', 'IGSF3', 'IL11', 'IL18', 'IL1R2', 'IMPA2', 'INPP5F', 'INSIG1', 'INSM1', 'IP6K1', 'IPCEF1', 'IRAK4', 'IRF1', 'IRF8', 'IRS2', 'IRX1', 'IRX2', 'IRX4', 'ISL1', 'ITGA11', 'ITGA3', 'ITGA9', 'ITGAV', 'ITGB4', 'ITGB6', 'ITGB8', 'ITIH5', 'ITPRIPL2', 'JAG1', 'JARID2', 'JRK', 'JSRP1', 'JUN', 'KANK1', 'KANK2', 'KCNA5', 'KCNAB1', 'KCNE3', 'KCNG1', 'KCNH6', 'KCNJ11', 'KCNJ16', 'KCNJ5', 'KCNJ6', 'KCNK16', 'KCNK3', 'KCNK5', 'KCNMA1', 'KCNN3', 'KCNQ1OT1', 'KCNQ1', 'KCTD12', 'KCTD8', 'KDELC2', 'KDELR3', 'KDM5D', 'KIF11', 'KIF12', 'KIF14', 'KIF21B', 'KIF2C', 'KIF5C', 'KISS1R', 'KLF2', 'KLF4', 'KLF5', 'KLF6', 'KLHDC8A', 'KLHL21', 'KLK11', 'KNSTRN', 'KPNA2', 'KRT17', 'KRTCAP3', 'L2HGDH', 'LAD1', 'LAMB1', 'LAMB2', 'LAPTM5', 'LBH', 'LDLR', 'LECT1', 'LEFTY1', 'LGALS3', 'LGALS9', 'LGI2', 'LHFP', 'LIMA1', 'LIMCH1', 'LIMK2', 'LIMS1', 'LIPH', 'LMO7', 'LONRF2', 'LOXL2', 'LPAR6', 'LRP12', 'LRP1', 'LRRC10B', 'LRRC59', 'LSAMP', 'LTBP4', 'LURAP1L', 'LUZP1', 'LXN', 'LY6E', 'LY6H', 'LYN', 'LYPD1', 'LYPD6B', 'MACC1', 'MAFA', 'MAFB', 'MAFF', 'MAL2', 'MALL', 'MAN1C1', 'MANF', 'MAOB', 'MAP1B', 'MAP2', 'MAP7D2', 'MAPT', 'MARCKSL1', 'MATN2', 'MDFI', 'MDK', 'MECOM', 'MEIS2', 'MERTK', 'METRN', 'METTL20', 'METTL22', 'MFI2', 'MGAT4A', 'MGAT4B', 'MGLL', 'MICAL2', 'MID1IP1', 'MIR143HG', 'MLLT11', 'MLXIPL', 'MMP14', 'MNX1', 'MPZL2', 'MRC1', 'MRC2', 'MSMO1', 'MSN', 'MSRB1', 'MSRB3', 'MTUS1', 'MUM1L1', 'MYC', 'MYH10', 'MYH9', 'MYL12A', 'MYO10', 'MYO1E', 'MYO5B', 'MYRF', 'MYZAP', 'NAA16', 'NCAM1', 'NCAPG', 'NCAPH', 'NCMAP', 'NDRG1', 'NDRG2', 'NDRG4', 'NDUFA4L2', 'NDUFA4', 'NEK8', 'NEURL1B', 'NEURL3', 'NEUROD1', 'NFATC4', 'NFKBIA', 'NFKBIE', 'NINJ1', 'NKD1', 'NKX2-2', 'NLN', 'NMB', 'NMNAT1', 'NOB1', 'NOTCH1', 'NOTCH2', 'NOTCH3', 'NOTCH4', 'NOX4', 'NPHS1', 'NPNT', 'NPTX2', 'NQO1', 'NR0B1', 'NR0B2', 'NR1D1', 'NR2F2', 'NR4A2', 'NR5A2', 'NRP2', 'NTN4', 'NUAK2', 'NUF2', 'NUSAP1', 'OCLN', 'OGDHL', 'OLFML2B', 'OPA3', 'ORM1', 'P4HA3', 'PACSIN2', 'PAH', 'PAK3', 'PALLD', 'PAMR1', 'PAPSS2', 'PARM1', 'PAX6', 'PBK', 'PCDH17', 'PCDH18', 'PCDH9', 'PCK1', 'PCNA', 'PCP4', 'PCSK2', 'PCSK5', 'PDE3A', 'PDE3B', 'PDE8B', 'PDGFA', 'PDGFD', 'PDIA4', 'PDLIM1', 'PDLIM4', 'PDLIM7', 'PDP2', 'PDX1', 'PEMT', 'PER3', 'PEX13', 'PFKFB3', 'PGBD5', 'PGM1', 'PGM2L1', 'PHF19', 'PHGDH', 'PHGR1', 'PHLDA2', 'PHLDB1', 'PID1', 'PIEZO1', 'PIK3AP1', 'PIK3R1', 'PIM1', 'PKIB', 'PKNOX2', 'PKP2', 'PLA2G4C', 'PLA2G7', 'PLAA', 'PLAGL1', 'PLAT', 'PLCXD3', 'PLEKHA2', 'PLEKHB1', 'PLEKHG2', 'PLEKHO1', 'PLIN5', 'PLK2', 'PLLP', 'PLOD2', 'PLSCR1', 'PLXDC1', 'PLXNA1', 'PLXNA2', 'PLXND1', 'PMAIP1', 'PNMA2', 'PNRC1', 'POC1A', 'POLD4', 'PON2', 'POU2F2', 'PPARGC1A', 'PPIC', 'PPIF', 'PPL', 'PPP1R15A', 'PPP1R18', 'PPP1R1A', 'PPP1R1B', 'PPP2R2B', 'PRC1', 'PRELP', 'PRKCDBP', 'PRKG1', 'PRKX', 'PRLHR', 'PRMT6', 'PRR11', 'PRR15L', 'PRSS22', 'PRSS23', 'PRSS8', 'PTAFR', 'PTGDS', 'PTGES', 'PTGFR', 'PTK7', 'PTPN14', 'PTPN3', 'PTPRN', 'PTPRT', 'PTRF', 'PTTG1', 'PVRL3', 'PVRL4', 'PYCARD', 'PYCR1', 'PYROXD2', 'QPRT', 'QSOX1', 'RAB11FIP1', 'RAB33A', 'RAB3B', 'RAB42', 'RACGAP1', 'RAMP1', 'RAP1GAP', 'RAPGEF5', 'RARG', 'RARRES1', 'RASA3', 'RASEF', 'RASGRP3', 'RASL12', 'RASSF4', 'RASSF9', 'RBM38', 'RBP1', 'RBPMS', 'RCL1', 'RCN3', 'RDH10', 'RDX', 'RELB', 'RETSAT', 'RFTN1', 'RFX6', 'RGS5', 'RHBDF1', 'RHOB', 'RHOC', 'RHOD', 'RHOV', 'RHPN2', 'RINL', 'RNASE6', 'RNF114', 'RNF157', 'RNF24', 'RNF32', 'ROBO1', 'ROR2', 'RPL22L1', 'RPS6KA2', 'RRAGD', 'RSU1', 'S100A10', 'S100A11', 'S100A16', 'S1PR3', 'SAMHD1', 'SCG2', 'SCG3', 'SCN1B', 'SCNN1A', 'SCTR', 'SDC2', 'SDS', 'SEL1L3', 'SEMA3B', 'SEMA3C', 'SEMA4B', 'SEMA6A', 'SEMA7A', 'SEPT3', 'SEPT6', 'SEPT9', 'SERPINA6', 'SERPINE2', 'SERPINF1', 'SERPINI1', 'SERTM1', 'SETX', 'SEZ6L', 'SFMBT2', 'SFN', 'SFRP1', 'SFRP5', 'SGOL1', 'SGSM1', 'SGSM3', 'SH3BGRL3', 'SH3BP4', 'SH3KBP1', 'SH3PXD2A', 'SHANK2', 'SHC1', 'SHROOM1', 'SHROOM3', 'SIRPA', 'SIX4', 'SKIL', 'SLC12A2', 'SLC12A7', 'SLC15A4', 'SLC16A3', 'SLC16A5', 'SLC16A7', 'SLC16A9', 'SLC1A5', 'SLC22A17', 'SLC22A3', 'SLC25A43', 'SLC26A4', 'SLC29A4', 'SLC2A13', 'SLC30A8', 'SLC38A1', 'SLC38A2', 'SLC38A5', 'SLC39A14', 'SLC39A5', 'SLC40A1', 'SLC43A3', 'SLC44A2', 'SLC45A3', 'SLC4A4', 'SLC4A5', 'SL
#> ℹ [2025-07-26 07:18:25] Data 3/5 of the srt_list has been log-normalized.
#> ℹ [2025-07-26 07:18:25] Perform FindVariableFeatures on the data 3/5 of the srt_list...
#> Warning: The following features were labelled as variable in 'var.features' but had no corresponding rank in `var.features.rank` and will therefore be ignored: 'ABCC3', 'ABCC8', 'ABCC9', 'ABCG1', 'ABCG2', 'ABHD2', 'ABHD4', 'ABI3', 'ACHE', 'ACP5', 'ACSL4', 'ACSS1', 'ACTN1', 'ACTN4', 'ADAM8', 'ADAM9', 'ADAMTS5', 'ADAMTS9', 'ADAMTSL2', 'ADAP2', 'ADCY1', 'ADCY3', 'ADCY5', 'ADM2', 'ADRA2A', 'ADRBK2', 'AEN', 'AGR2', 'AGRN', 'AGTRAP', 'AHNAK', 'AHR', 'AKAP7', 'ALDH1A1', 'ALDH1A2', 'ALDH1B1', 'ALOX5AP', 'ALS2CL', 'AMOTL2', 'AMT', 'ANK1', 'ANKRD1', 'ANKRD37', 'ANLN', 'ANO6', 'ANXA13', 'ANXA3', 'ANXA9', 'AP5Z1', 'AQP3', 'ARC', 'ARG2', 'ARHGAP22', 'ARHGAP23', 'ARHGAP29', 'ARHGEF10L', 'ARHGEF2', 'ARHGEF39', 'ARHGEF40', 'ARL14', 'ARL4C', 'ARL6IP1', 'ARNTL2', 'ARX', 'ASAP1', 'ASAP2', 'ASCL2', 'ASF1B', 'ASGR1', 'ASNS', 'ASRGL1', 'ASS1', 'ATF3', 'ATF5', 'ATP11A', 'ATP1A1', 'ATP1B1', 'ATP1B2', 'ATP2A3', 'ATP8A1', 'B4GALT1', 'B4GALT5', 'BACE2', 'BAIAP2L1', 'BAIAP3', 'BAMBI', 'BBC3', 'BCL2L15', 'BCL3', 'BHLHE40', 'BHLHE41', 'BIK', 'BIRC5', 'BLNK', 'BMF', 'BMP1', 'BMP2', 'BMP7', 'BST2', 'BUB1B', 'BUB1', 'C1QA', 'C1QC', 'C1QL1', 'C2CD4A', 'C2CD4B', 'C2', 'CACNA1A', 'CACNA2D1', 'CACNG4', 'CADM2', 'CADPS', 'CALY', 'CAMK2B', 'CAPG', 'CAPN5', 'CAPN6', 'CARTPT', 'CASP9', 'CAV2', 'CBLB', 'CBLN4', 'CBS', 'CCBE1', 'CCDC34', 'CCDC51', 'CCDC69', 'CCNA1', 'CD14', 'CD276', 'CD300A', 'CD68', 'CD82', 'CD9', 'CDC20', 'CDC42EP3', 'CDCA8', 'CDH17', 'CDH3', 'CDK1', 'CDK5R2', 'CDKN1A', 'CDKN2A', 'CDKN2B', 'CDKN3', 'CDR2L', 'CDX2', 'CEBPB', 'CELF3', 'CENPW', 'CERCAM', 'CFI', 'CHAC1', 'CHDH', 'CHGB', 'CHRNA3', 'CITED2', 'CITED4', 'CKAP2', 'CKAP5', 'CKMT2', 'CKS1B', 'CKS2', 'CLCF1', 'CLDN1', 'CLIC4', 'CLMN', 'CLSPN', 'CMPK2', 'CMTM7', 'CNIH2', 'CNKSR2', 'CNN2', 'CNN3', 'CNTN1', 'COL13A1', 'COL16A1', 'COL18A1', 'COL4A2', 'COL5A3', 'COL9A2', 'COPG1', 'COPZ2', 'CORO1C', 'COTL1', 'COX7A1', 'CPLX2', 'CPPED1', 'CPVL', 'CRABP2', 'CRAT', 'CREB3L1', 'CREB5', 'CRIP1', 'CRLF1', 'CRTAC1', 'CRTAP', 'CRYM', 'CSF1R', 'CSGALNACT1', 'CTSC', 'CTSH', 'CTSO', 'CTSZ', 'CWC25', 'CX3CL1', 'CXADR', 'CXCL12', 'CXCL16', 'CXCL1', 'CYB5A', 'CYP26B1', 'CYP27A1', 'CYTH3', 'DAB2IP', 'DACH1', 'DACT1', 'DAPP1', 'DBN1', 'DBNDD1', 'DCUN1D2', 'DDC', 'DDIT4', 'DDX51', 'DEDD2', 'DHODH', 'DHRS2', 'DIP2A', 'DIRAS2', 'DNAJC12', 'DNAL1', 'DOCK4', 'DOCK6', 'DOCK8', 'DOCK9', 'DOK5', 'DPP4', 'DPP6', 'DSC2', 'DSEL', 'DUSP1', 'DUSP23', 'DUSP26', 'DUSP5', 'ECEL1', 'ECM1', 'ECT2', 'EDN2', 'EEF1D', 'EEF2K', 'EFEMP2', 'EFHD2', 'EFNA1', 'EGFR', 'EGR1', 'EGR4', 'EHD2', 'EHD4', 'EHF', 'ELAVL4', 'EMILIN1', 'EMP3', 'ENAH', 'ENO2', 'ENPP2', 'EPHB2', 'EPHB4', 'EPS8', 'ERBB3', 'ERO1L', 'ERRFI1', 'ETS2', 'EVA1A', 'EVA1B', 'F2RL2', 'F2RL3', 'F2R', 'F5', 'FA2H', 'FAM105A', 'FAM107B', 'FAM129B', 'FAM134B', 'FAM171A1', 'FAM227A', 'FAM43A', 'FAM46A', 'FAM46C', 'FAM84A', 'FBLIM1', 'FBP1', 'FBXL20', 'FBXO2', 'FBXO32', 'FBXO5', 'FBXO6', 'FCER1G', 'FCGRT', 'FERMT1', 'FEV', 'FFAR4', 'FGB', 'FGF13', 'FGFR2', 'FGFR3', 'FHL2', 'FHOD1', 'FJX1', 'FKBP10', 'FKBP11', 'FKBP5', 'FLNA', 'FMNL3', 'FNDC4', 'FNIP2', 'FOSB', 'FOSL2', 'FOXA3', 'FOXC1', 'FOXM1', 'FOXQ1', 'FRZB', 'FSCN1', 'FSTL3', 'FTH1', 'FURIN', 'FUT8', 'FXYD3', 'FXYD5', 'FXYD6', 'G0S2', 'G6PC', 'GAD2', 'GADD45A', 'GADD45B', 'GALE', 'GALM', 'GALNT2', 'GAL', 'GAS6', 'GAS7', 'GATA2', 'GATA4', 'GATSL3', 'GCGR', 'GCH1', 'GCK', 'GCLC', 'GCNT1', 'GCNT3', 'GC', 'GDA', 'GEM', 'GFOD2', 'GFPT2', 'GFRA1', 'GJA1', 'GJB1', 'GK', 'GLIPR1', 'GLRX', 'GLS', 'GMNN', 'GNPAT', 'GOLM1', 'GPC1', 'GPC6', 'GPD1', 'GPR160', 'GPR161', 'GPSM1', 'GPX3', 'GPX8', 'GRASP', 'GREB1', 'GSN', 'GSTM5', 'GSTP1', 'GTF3C3', 'GTPBP10', 'GTSE1', 'GULP1', 'H19', 'HABP2', 'HAND2', 'HDAC9', 'HEPACAM2', 'HEPH', 'HERPUD1', 'HES1', 'HEYL', 'HHEX', 'HIF1A', 'HIF3A', 'HILPDA', 'HIST1H1C', 'HIST1H2BK', 'HIST2H2BE', 'HK1', 'HK2', 'HKDC1', 'HMGB2', 'HMGB3', 'HMGCR', 'HMGCS1', 'HMMR', 'HN1', 'HNF1B', 'HOGA1', 'HOMER3', 'HOPX', 'HOXB2', 'HPCAL1', 'HPGD', 'HPN', 'HS3ST1', 'HSD11B2', 'HSD17B11', 'HSP90B1', 'HSPB1', 'HSPB8', 'HSPH1', 'ID1', 'ID4', 'IDH1', 'IDH2', 'IER2', 'IER3', 'IFIT1', 'IFITM2', 'IFNLR1', 'IGSF10', 'IGSF1', 'IGSF3', 'IL11', 'IL18', 'IL1R2', 'IMPA2', 'INPP5F', 'INSIG1', 'INSM1', 'IP6K1', 'IPCEF1', 'IRAK4', 'IRF1', 'IRF8', 'IRS2', 'IRX1', 'IRX2', 'IRX4', 'ISL1', 'ITGA11', 'ITGA3', 'ITGA9', 'ITGAV', 'ITGB4', 'ITGB6', 'ITGB8', 'ITIH5', 'ITPRIPL2', 'JAG1', 'JARID2', 'JRK', 'JSRP1', 'JUN', 'KANK1', 'KANK2', 'KCNA5', 'KCNAB1', 'KCNE3', 'KCNG1', 'KCNH6', 'KCNJ11', 'KCNJ16', 'KCNJ5', 'KCNJ6', 'KCNK16', 'KCNK3', 'KCNK5', 'KCNMA1', 'KCNN3', 'KCNQ1OT1', 'KCNQ1', 'KCTD12', 'KCTD8', 'KDELC2', 'KDELR3', 'KDM5D', 'KIF11', 'KIF12', 'KIF14', 'KIF21B', 'KIF2C', 'KIF5C', 'KISS1R', 'KLF2', 'KLF4', 'KLF5', 'KLF6', 'KLHDC8A', 'KLHL21', 'KLK11', 'KNSTRN', 'KPNA2', 'KRT17', 'KRTCAP3', 'L2HGDH', 'LAD1', 'LAMB1', 'LAMB2', 'LAPTM5', 'LBH', 'LDLR', 'LECT1', 'LEFTY1', 'LGALS3', 'LGALS9', 'LGI2', 'LHFP', 'LIMA1', 'LIMCH1', 'LIMK2', 'LIMS1', 'LIPH', 'LMO7', 'LONRF2', 'LOXL2', 'LPAR6', 'LRP12', 'LRP1', 'LRRC10B', 'LRRC59', 'LSAMP', 'LTBP4', 'LURAP1L', 'LUZP1', 'LXN', 'LY6E', 'LY6H', 'LYN', 'LYPD1', 'LYPD6B', 'MACC1', 'MAFA', 'MAFB', 'MAFF', 'MAL2', 'MALL', 'MAN1C1', 'MANF', 'MAOB', 'MAP1B', 'MAP2', 'MAP7D2', 'MAPT', 'MARCKSL1', 'MATN2', 'MDFI', 'MDK', 'MECOM', 'MEIS2', 'MERTK', 'METRN', 'METTL20', 'METTL22', 'MFI2', 'MGAT4A', 'MGAT4B', 'MGLL', 'MICAL2', 'MID1IP1', 'MIR143HG', 'MLLT11', 'MLXIPL', 'MMP14', 'MNX1', 'MPZL2', 'MRC1', 'MRC2', 'MSMO1', 'MSN', 'MSRB1', 'MSRB3', 'MTUS1', 'MUM1L1', 'MYC', 'MYH10', 'MYH9', 'MYL12A', 'MYO10', 'MYO1E', 'MYO5B', 'MYRF', 'MYZAP', 'NAA16', 'NCAM1', 'NCAPG', 'NCAPH', 'NCMAP', 'NDRG1', 'NDRG2', 'NDRG4', 'NDUFA4L2', 'NDUFA4', 'NEK8', 'NEURL1B', 'NEURL3', 'NEUROD1', 'NFATC4', 'NFKBIA', 'NFKBIE', 'NINJ1', 'NKD1', 'NKX2-2', 'NLN', 'NMB', 'NMNAT1', 'NOB1', 'NOTCH1', 'NOTCH2', 'NOTCH3', 'NOTCH4', 'NOX4', 'NPHS1', 'NPNT', 'NPTX2', 'NQO1', 'NR0B1', 'NR0B2', 'NR1D1', 'NR2F2', 'NR4A2', 'NR5A2', 'NRP2', 'NTN4', 'NUAK2', 'NUF2', 'NUSAP1', 'OCLN', 'OGDHL', 'OLFML2B', 'OPA3', 'ORM1', 'P4HA3', 'PACSIN2', 'PAH', 'PAK3', 'PALLD', 'PAMR1', 'PAPSS2', 'PARM1', 'PAX6', 'PBK', 'PCDH17', 'PCDH18', 'PCDH9', 'PCK1', 'PCNA', 'PCP4', 'PCSK2', 'PCSK5', 'PDE3A', 'PDE3B', 'PDE8B', 'PDGFA', 'PDGFD', 'PDIA4', 'PDLIM1', 'PDLIM4', 'PDLIM7', 'PDP2', 'PDX1', 'PEMT', 'PER3', 'PEX13', 'PFKFB3', 'PGBD5', 'PGM1', 'PGM2L1', 'PHF19', 'PHGDH', 'PHGR1', 'PHLDA2', 'PHLDB1', 'PID1', 'PIEZO1', 'PIK3AP1', 'PIK3R1', 'PIM1', 'PKIB', 'PKNOX2', 'PKP2', 'PLA2G4C', 'PLA2G7', 'PLAA', 'PLAGL1', 'PLAT', 'PLCXD3', 'PLEKHA2', 'PLEKHB1', 'PLEKHG2', 'PLEKHO1', 'PLIN5', 'PLK2', 'PLLP', 'PLOD2', 'PLSCR1', 'PLXDC1', 'PLXNA1', 'PLXNA2', 'PLXND1', 'PMAIP1', 'PNMA2', 'PNRC1', 'POC1A', 'POLD4', 'PON2', 'POU2F2', 'PPARGC1A', 'PPIC', 'PPIF', 'PPL', 'PPP1R15A', 'PPP1R18', 'PPP1R1A', 'PPP1R1B', 'PPP2R2B', 'PRC1', 'PRELP', 'PRKCDBP', 'PRKG1', 'PRKX', 'PRLHR', 'PRMT6', 'PRR11', 'PRR15L', 'PRSS22', 'PRSS23', 'PRSS8', 'PTAFR', 'PTGDS', 'PTGES', 'PTGFR', 'PTK7', 'PTPN14', 'PTPN3', 'PTPRN', 'PTPRT', 'PTRF', 'PTTG1', 'PVRL3', 'PVRL4', 'PYCARD', 'PYCR1', 'PYROXD2', 'QPRT', 'QSOX1', 'RAB11FIP1', 'RAB33A', 'RAB3B', 'RAB42', 'RACGAP1', 'RAMP1', 'RAP1GAP', 'RAPGEF5', 'RARG', 'RARRES1', 'RASA3', 'RASEF', 'RASGRP3', 'RASL12', 'RASSF4', 'RASSF9', 'RBM38', 'RBP1', 'RBPMS', 'RCL1', 'RCN3', 'RDH10', 'RDX', 'RELB', 'RETSAT', 'RFTN1', 'RFX6', 'RGS5', 'RHBDF1', 'RHOB', 'RHOC', 'RHOD', 'RHOV', 'RHPN2', 'RINL', 'RNASE6', 'RNF114', 'RNF157', 'RNF24', 'RNF32', 'ROBO1', 'ROR2', 'RPL22L1', 'RPS6KA2', 'RRAGD', 'RSU1', 'S100A10', 'S100A11', 'S100A16', 'S1PR3', 'SAMHD1', 'SCG2', 'SCG3', 'SCN1B', 'SCNN1A', 'SCTR', 'SDC2', 'SDS', 'SEL1L3', 'SEMA3B', 'SEMA3C', 'SEMA4B', 'SEMA6A', 'SEMA7A', 'SEPT3', 'SEPT6', 'SEPT9', 'SERPINA6', 'SERPINE2', 'SERPINF1', 'SERPINI1', 'SERTM1', 'SETX', 'SEZ6L', 'SFMBT2', 'SFN', 'SFRP1', 'SFRP5', 'SGOL1', 'SGSM1', 'SGSM3', 'SH3BGRL3', 'SH3BP4', 'SH3KBP1', 'SH3PXD2A', 'SHANK2', 'SHC1', 'SHROOM1', 'SHROOM3', 'SIRPA', 'SIX4', 'SKIL', 'SLC12A2', 'SLC12A7', 'SLC15A4', 'SLC16A3', 'SLC16A5', 'SLC16A7', 'SLC16A9', 'SLC1A5', 'SLC22A17', 'SLC22A3', 'SLC25A43', 'SLC26A4', 'SLC29A4', 'SLC2A13', 'SLC30A8', 'SLC38A1', 'SLC38A2', 'SLC38A5', 'SLC39A14', 'SLC39A5', 'SLC40A1', 'SLC43A3', 'SLC44A2', 'SLC45A3', 'SLC4A4', 'SLC4A5', 'SL
#> ℹ [2025-07-26 07:18:26] Data 4/5 of the srt_list has been log-normalized.
#> ℹ [2025-07-26 07:18:26] Perform FindVariableFeatures on the data 4/5 of the srt_list...
#> Warning: The following features were labelled as variable in 'var.features' but had no corresponding rank in `var.features.rank` and will therefore be ignored: 'ABCC3', 'ABCC8', 'ABCC9', 'ABCG1', 'ABCG2', 'ABHD2', 'ABHD4', 'ABI3', 'ACHE', 'ACP5', 'ACSL4', 'ACSS1', 'ACTN1', 'ACTN4', 'ADAM8', 'ADAM9', 'ADAMTS5', 'ADAMTS9', 'ADAMTSL2', 'ADAP2', 'ADCY1', 'ADCY3', 'ADCY5', 'ADM2', 'ADRA2A', 'ADRBK2', 'AEN', 'AGR2', 'AGRN', 'AGTRAP', 'AHNAK', 'AHR', 'AKAP7', 'ALDH1A1', 'ALDH1A2', 'ALDH1B1', 'ALOX5AP', 'ALS2CL', 'AMOTL2', 'AMT', 'ANK1', 'ANKRD1', 'ANKRD37', 'ANLN', 'ANO6', 'ANXA13', 'ANXA3', 'ANXA9', 'AP5Z1', 'AQP3', 'ARC', 'ARG2', 'ARHGAP22', 'ARHGAP23', 'ARHGAP29', 'ARHGEF10L', 'ARHGEF2', 'ARHGEF39', 'ARHGEF40', 'ARL14', 'ARL4C', 'ARL6IP1', 'ARNTL2', 'ARX', 'ASAP1', 'ASAP2', 'ASCL2', 'ASF1B', 'ASGR1', 'ASNS', 'ASRGL1', 'ASS1', 'ATF3', 'ATF5', 'ATP11A', 'ATP1A1', 'ATP1B1', 'ATP1B2', 'ATP2A3', 'ATP8A1', 'B4GALT1', 'B4GALT5', 'BACE2', 'BAIAP2L1', 'BAIAP3', 'BAMBI', 'BBC3', 'BCL2L15', 'BCL3', 'BHLHE40', 'BHLHE41', 'BIK', 'BIRC5', 'BLNK', 'BMF', 'BMP1', 'BMP2', 'BMP7', 'BST2', 'BUB1B', 'BUB1', 'C1QA', 'C1QC', 'C1QL1', 'C2CD4A', 'C2CD4B', 'C2', 'CACNA1A', 'CACNA2D1', 'CACNG4', 'CADM2', 'CADPS', 'CALY', 'CAMK2B', 'CAPG', 'CAPN5', 'CAPN6', 'CARTPT', 'CASP9', 'CAV2', 'CBLB', 'CBLN4', 'CBS', 'CCBE1', 'CCDC34', 'CCDC51', 'CCDC69', 'CCNA1', 'CD14', 'CD276', 'CD300A', 'CD68', 'CD82', 'CD9', 'CDC20', 'CDC42EP3', 'CDCA8', 'CDH17', 'CDH3', 'CDK1', 'CDK5R2', 'CDKN1A', 'CDKN2A', 'CDKN2B', 'CDKN3', 'CDR2L', 'CDX2', 'CEBPB', 'CELF3', 'CENPW', 'CERCAM', 'CFI', 'CHAC1', 'CHDH', 'CHGB', 'CHRNA3', 'CITED2', 'CITED4', 'CKAP2', 'CKAP5', 'CKMT2', 'CKS1B', 'CKS2', 'CLCF1', 'CLDN1', 'CLIC4', 'CLMN', 'CLSPN', 'CMPK2', 'CMTM7', 'CNIH2', 'CNKSR2', 'CNN2', 'CNN3', 'CNTN1', 'COL13A1', 'COL16A1', 'COL18A1', 'COL4A2', 'COL5A3', 'COL9A2', 'COPG1', 'COPZ2', 'CORO1C', 'COTL1', 'COX7A1', 'CPLX2', 'CPPED1', 'CPVL', 'CRABP2', 'CRAT', 'CREB3L1', 'CREB5', 'CRIP1', 'CRLF1', 'CRTAC1', 'CRTAP', 'CRYM', 'CSF1R', 'CSGALNACT1', 'CTSC', 'CTSH', 'CTSO', 'CTSZ', 'CWC25', 'CX3CL1', 'CXADR', 'CXCL12', 'CXCL16', 'CXCL1', 'CYB5A', 'CYP26B1', 'CYP27A1', 'CYTH3', 'DAB2IP', 'DACH1', 'DACT1', 'DAPP1', 'DBN1', 'DBNDD1', 'DCUN1D2', 'DDC', 'DDIT4', 'DDX51', 'DEDD2', 'DHODH', 'DHRS2', 'DIP2A', 'DIRAS2', 'DNAJC12', 'DNAL1', 'DOCK4', 'DOCK6', 'DOCK8', 'DOCK9', 'DOK5', 'DPP4', 'DPP6', 'DSC2', 'DSEL', 'DUSP1', 'DUSP23', 'DUSP26', 'DUSP5', 'ECEL1', 'ECM1', 'ECT2', 'EDN2', 'EEF1D', 'EEF2K', 'EFEMP2', 'EFHD2', 'EFNA1', 'EGFR', 'EGR1', 'EGR4', 'EHD2', 'EHD4', 'EHF', 'ELAVL4', 'EMILIN1', 'EMP3', 'ENAH', 'ENO2', 'ENPP2', 'EPHB2', 'EPHB4', 'EPS8', 'ERBB3', 'ERO1L', 'ERRFI1', 'ETS2', 'EVA1A', 'EVA1B', 'F2RL2', 'F2RL3', 'F2R', 'F5', 'FA2H', 'FAM105A', 'FAM107B', 'FAM129B', 'FAM134B', 'FAM171A1', 'FAM227A', 'FAM43A', 'FAM46A', 'FAM46C', 'FAM84A', 'FBLIM1', 'FBP1', 'FBXL20', 'FBXO2', 'FBXO32', 'FBXO5', 'FBXO6', 'FCER1G', 'FCGRT', 'FERMT1', 'FEV', 'FFAR4', 'FGB', 'FGF13', 'FGFR2', 'FGFR3', 'FHL2', 'FHOD1', 'FJX1', 'FKBP10', 'FKBP11', 'FKBP5', 'FLNA', 'FMNL3', 'FNDC4', 'FNIP2', 'FOSB', 'FOSL2', 'FOXA3', 'FOXC1', 'FOXM1', 'FOXQ1', 'FRZB', 'FSCN1', 'FSTL3', 'FTH1', 'FURIN', 'FUT8', 'FXYD3', 'FXYD5', 'FXYD6', 'G0S2', 'G6PC', 'GAD2', 'GADD45A', 'GADD45B', 'GALE', 'GALM', 'GALNT2', 'GAL', 'GAS6', 'GAS7', 'GATA2', 'GATA4', 'GATSL3', 'GCGR', 'GCH1', 'GCK', 'GCLC', 'GCNT1', 'GCNT3', 'GC', 'GDA', 'GEM', 'GFOD2', 'GFPT2', 'GFRA1', 'GJA1', 'GJB1', 'GK', 'GLIPR1', 'GLRX', 'GLS', 'GMNN', 'GNPAT', 'GOLM1', 'GPC1', 'GPC6', 'GPD1', 'GPR160', 'GPR161', 'GPSM1', 'GPX3', 'GPX8', 'GRASP', 'GREB1', 'GSN', 'GSTM5', 'GSTP1', 'GTF3C3', 'GTPBP10', 'GTSE1', 'GULP1', 'H19', 'HABP2', 'HAND2', 'HDAC9', 'HEPACAM2', 'HEPH', 'HERPUD1', 'HES1', 'HEYL', 'HHEX', 'HIF1A', 'HIF3A', 'HILPDA', 'HIST1H1C', 'HIST1H2BK', 'HIST2H2BE', 'HK1', 'HK2', 'HKDC1', 'HMGB2', 'HMGB3', 'HMGCR', 'HMGCS1', 'HMMR', 'HN1', 'HNF1B', 'HOGA1', 'HOMER3', 'HOPX', 'HOXB2', 'HPCAL1', 'HPGD', 'HPN', 'HS3ST1', 'HSD11B2', 'HSD17B11', 'HSP90B1', 'HSPB1', 'HSPB8', 'HSPH1', 'ID1', 'ID4', 'IDH1', 'IDH2', 'IER2', 'IER3', 'IFIT1', 'IFITM2', 'IFNLR1', 'IGSF10', 'IGSF1', 'IGSF3', 'IL11', 'IL18', 'IL1R2', 'IMPA2', 'INPP5F', 'INSIG1', 'INSM1', 'IP6K1', 'IPCEF1', 'IRAK4', 'IRF1', 'IRF8', 'IRS2', 'IRX1', 'IRX2', 'IRX4', 'ISL1', 'ITGA11', 'ITGA3', 'ITGA9', 'ITGAV', 'ITGB4', 'ITGB6', 'ITGB8', 'ITIH5', 'ITPRIPL2', 'JAG1', 'JARID2', 'JRK', 'JSRP1', 'JUN', 'KANK1', 'KANK2', 'KCNA5', 'KCNAB1', 'KCNE3', 'KCNG1', 'KCNH6', 'KCNJ11', 'KCNJ16', 'KCNJ5', 'KCNJ6', 'KCNK16', 'KCNK3', 'KCNK5', 'KCNMA1', 'KCNN3', 'KCNQ1OT1', 'KCNQ1', 'KCTD12', 'KCTD8', 'KDELC2', 'KDELR3', 'KDM5D', 'KIF11', 'KIF12', 'KIF14', 'KIF21B', 'KIF2C', 'KIF5C', 'KISS1R', 'KLF2', 'KLF4', 'KLF5', 'KLF6', 'KLHDC8A', 'KLHL21', 'KLK11', 'KNSTRN', 'KPNA2', 'KRT17', 'KRTCAP3', 'L2HGDH', 'LAD1', 'LAMB1', 'LAMB2', 'LAPTM5', 'LBH', 'LDLR', 'LECT1', 'LEFTY1', 'LGALS3', 'LGALS9', 'LGI2', 'LHFP', 'LIMA1', 'LIMCH1', 'LIMK2', 'LIMS1', 'LIPH', 'LMO7', 'LONRF2', 'LOXL2', 'LPAR6', 'LRP12', 'LRP1', 'LRRC10B', 'LRRC59', 'LSAMP', 'LTBP4', 'LURAP1L', 'LUZP1', 'LXN', 'LY6E', 'LY6H', 'LYN', 'LYPD1', 'LYPD6B', 'MACC1', 'MAFA', 'MAFB', 'MAFF', 'MAL2', 'MALL', 'MAN1C1', 'MANF', 'MAOB', 'MAP1B', 'MAP2', 'MAP7D2', 'MAPT', 'MARCKSL1', 'MATN2', 'MDFI', 'MDK', 'MECOM', 'MEIS2', 'MERTK', 'METRN', 'METTL20', 'METTL22', 'MFI2', 'MGAT4A', 'MGAT4B', 'MGLL', 'MICAL2', 'MID1IP1', 'MIR143HG', 'MLLT11', 'MLXIPL', 'MMP14', 'MNX1', 'MPZL2', 'MRC1', 'MRC2', 'MSMO1', 'MSN', 'MSRB1', 'MSRB3', 'MTUS1', 'MUM1L1', 'MYC', 'MYH10', 'MYH9', 'MYL12A', 'MYO10', 'MYO1E', 'MYO5B', 'MYRF', 'MYZAP', 'NAA16', 'NCAM1', 'NCAPG', 'NCAPH', 'NCMAP', 'NDRG1', 'NDRG2', 'NDRG4', 'NDUFA4L2', 'NDUFA4', 'NEK8', 'NEURL1B', 'NEURL3', 'NEUROD1', 'NFATC4', 'NFKBIA', 'NFKBIE', 'NINJ1', 'NKD1', 'NKX2-2', 'NLN', 'NMB', 'NMNAT1', 'NOB1', 'NOTCH1', 'NOTCH2', 'NOTCH3', 'NOTCH4', 'NOX4', 'NPHS1', 'NPNT', 'NPTX2', 'NQO1', 'NR0B1', 'NR0B2', 'NR1D1', 'NR2F2', 'NR4A2', 'NR5A2', 'NRP2', 'NTN4', 'NUAK2', 'NUF2', 'NUSAP1', 'OCLN', 'OGDHL', 'OLFML2B', 'OPA3', 'ORM1', 'P4HA3', 'PACSIN2', 'PAH', 'PAK3', 'PALLD', 'PAMR1', 'PAPSS2', 'PARM1', 'PAX6', 'PBK', 'PCDH17', 'PCDH18', 'PCDH9', 'PCK1', 'PCNA', 'PCP4', 'PCSK2', 'PCSK5', 'PDE3A', 'PDE3B', 'PDE8B', 'PDGFA', 'PDGFD', 'PDIA4', 'PDLIM1', 'PDLIM4', 'PDLIM7', 'PDP2', 'PDX1', 'PEMT', 'PER3', 'PEX13', 'PFKFB3', 'PGBD5', 'PGM1', 'PGM2L1', 'PHF19', 'PHGDH', 'PHGR1', 'PHLDA2', 'PHLDB1', 'PID1', 'PIEZO1', 'PIK3AP1', 'PIK3R1', 'PIM1', 'PKIB', 'PKNOX2', 'PKP2', 'PLA2G4C', 'PLA2G7', 'PLAA', 'PLAGL1', 'PLAT', 'PLCXD3', 'PLEKHA2', 'PLEKHB1', 'PLEKHG2', 'PLEKHO1', 'PLIN5', 'PLK2', 'PLLP', 'PLOD2', 'PLSCR1', 'PLXDC1', 'PLXNA1', 'PLXNA2', 'PLXND1', 'PMAIP1', 'PNMA2', 'PNRC1', 'POC1A', 'POLD4', 'PON2', 'POU2F2', 'PPARGC1A', 'PPIC', 'PPIF', 'PPL', 'PPP1R15A', 'PPP1R18', 'PPP1R1A', 'PPP1R1B', 'PPP2R2B', 'PRC1', 'PRELP', 'PRKCDBP', 'PRKG1', 'PRKX', 'PRLHR', 'PRMT6', 'PRR11', 'PRR15L', 'PRSS22', 'PRSS23', 'PRSS8', 'PTAFR', 'PTGDS', 'PTGES', 'PTGFR', 'PTK7', 'PTPN14', 'PTPN3', 'PTPRN', 'PTPRT', 'PTRF', 'PTTG1', 'PVRL3', 'PVRL4', 'PYCARD', 'PYCR1', 'PYROXD2', 'QPRT', 'QSOX1', 'RAB11FIP1', 'RAB33A', 'RAB3B', 'RAB42', 'RACGAP1', 'RAMP1', 'RAP1GAP', 'RAPGEF5', 'RARG', 'RARRES1', 'RASA3', 'RASEF', 'RASGRP3', 'RASL12', 'RASSF4', 'RASSF9', 'RBM38', 'RBP1', 'RBPMS', 'RCL1', 'RCN3', 'RDH10', 'RDX', 'RELB', 'RETSAT', 'RFTN1', 'RFX6', 'RGS5', 'RHBDF1', 'RHOB', 'RHOC', 'RHOD', 'RHOV', 'RHPN2', 'RINL', 'RNASE6', 'RNF114', 'RNF157', 'RNF24', 'RNF32', 'ROBO1', 'ROR2', 'RPL22L1', 'RPS6KA2', 'RRAGD', 'RSU1', 'S100A10', 'S100A11', 'S100A16', 'S1PR3', 'SAMHD1', 'SCG2', 'SCG3', 'SCN1B', 'SCNN1A', 'SCTR', 'SDC2', 'SDS', 'SEL1L3', 'SEMA3B', 'SEMA3C', 'SEMA4B', 'SEMA6A', 'SEMA7A', 'SEPT3', 'SEPT6', 'SEPT9', 'SERPINA6', 'SERPINE2', 'SERPINF1', 'SERPINI1', 'SERTM1', 'SETX', 'SEZ6L', 'SFMBT2', 'SFN', 'SFRP1', 'SFRP5', 'SGOL1', 'SGSM1', 'SGSM3', 'SH3BGRL3', 'SH3BP4', 'SH3KBP1', 'SH3PXD2A', 'SHANK2', 'SHC1', 'SHROOM1', 'SHROOM3', 'SIRPA', 'SIX4', 'SKIL', 'SLC12A2', 'SLC12A7', 'SLC15A4', 'SLC16A3', 'SLC16A5', 'SLC16A7', 'SLC16A9', 'SLC1A5', 'SLC22A17', 'SLC22A3', 'SLC25A43', 'SLC26A4', 'SLC29A4', 'SLC2A13', 'SLC30A8', 'SLC38A1', 'SLC38A2', 'SLC38A5', 'SLC39A14', 'SLC39A5', 'SLC40A1', 'SLC43A3', 'SLC44A2', 'SLC45A3', 'SLC4A4', 'SLC4A5', 'SL
#> ℹ [2025-07-26 07:18:26] Data 5/5 of the srt_list has been log-normalized.
#> ℹ [2025-07-26 07:18:26] Perform FindVariableFeatures on the data 5/5 of the srt_list...
#> ℹ [2025-07-26 07:18:27] Use the separate HVF from srt_list...
#> ℹ [2025-07-26 07:18:27] Number of available HVF: 259
#> ℹ [2025-07-26 07:18:27] Finished checking.
#> ℹ [2025-07-26 07:18:29] Perform integration(Uncorrected) on the data...
#> ℹ [2025-07-26 07:18:29] Perform ScaleData on the data...
#> ℹ [2025-07-26 07:18:29] Perform linear dimension reduction (pca) on the data...
#> ℹ [2025-07-26 07:18:29] Linear_reduction(pca) is already existed. Skip calculation.
#> Warning: The following arguments are not used: features
#> Warning: multiple layers are identified by counts.1.1.1.1 counts.2.1 counts.2.1.1 counts.2.1.1.1 data.1.1.1.1 scale.data.1.1.1.1 data.2.1.1.1 scale.data.2.1.1.1 data.2.1.1 scale.data.2.1.1 data.2.1 scale.data.2.1
#> only the first layer is used
#> ! [2025-07-26 07:18:29] Error: Not compatible with requested type: [type=S4; target=double].
#> ! [2025-07-26 07:18:29] Error when performing FindClusters. Skip this step...
#> ℹ [2025-07-26 07:18:29] Perform nonlinear dimension reduction (umap) on the data...
#> Warning: no non-missing arguments to min; returning Inf
#> Warning: no non-missing arguments to max; returning -Inf
#> ℹ [2025-07-26 07:18:29] Non-linear dimensionality reduction(umap) using Reduction(Uncorrectedpca, dims:Inf--Inf) as input
#> ! [2025-07-26 07:18:29] Error in `object[[graph]]`:
#> ! [2025-07-26 07:18:29] ! ‘Uncorrected_SNN’ not found in this Seurat object
#> ! [2025-07-26 07:18:29]
#> ! [2025-07-26 07:18:29] Error when performing nonlinear dimension reduction. Skip this step...
#> ℹ [2025-07-26 07:18:31] Uncorrected_integrate done
#> ℹ [2025-07-26 07:18:31] Elapsed time: 8.71 secs
CellDimPlot(
panc8_sub,
group.by = c("tech", "celltype")
)
#> Warning: No shared levels found between `names(values)` of the manual scale and the
#> data's fill values.
#> Warning: No shared levels found between `names(values)` of the manual scale and the
#> data's fill values.
panc8_sub <- integration_scop(
panc8_sub,
batch = "tech",
integration_method = "Uncorrected",
HVF_min_intersection = 5
)
#> ℹ [2025-07-26 07:18:22] Start Uncorrected_integrate
#> ℹ [2025-07-26 07:18:22] Spliting srt_merge into srt_list by column tech...
#> ℹ [2025-07-26 07:18:24] Checking srt_list...
#> Warning: The following features were labelled as variable in 'var.features' but had no corresponding rank in `var.features.rank` and will therefore be ignored: 'ABCC3', 'ABCC8', 'ABCC9', 'ABCG1', 'ABCG2', 'ABHD2', 'ABHD4', 'ABI3', 'ACHE', 'ACP5', 'ACSL4', 'ACSS1', 'ACTN1', 'ACTN4', 'ADAM8', 'ADAM9', 'ADAMTS5', 'ADAMTS9', 'ADAMTSL2', 'ADAP2', 'ADCY1', 'ADCY3', 'ADCY5', 'ADM2', 'ADRA2A', 'ADRBK2', 'AEN', 'AGR2', 'AGRN', 'AGTRAP', 'AHNAK', 'AHR', 'AKAP7', 'ALDH1A1', 'ALDH1A2', 'ALDH1B1', 'ALOX5AP', 'ALS2CL', 'AMOTL2', 'AMT', 'ANK1', 'ANKRD1', 'ANKRD37', 'ANLN', 'ANO6', 'ANXA13', 'ANXA3', 'ANXA9', 'AP5Z1', 'AQP3', 'ARC', 'ARG2', 'ARHGAP22', 'ARHGAP23', 'ARHGAP29', 'ARHGEF10L', 'ARHGEF2', 'ARHGEF39', 'ARHGEF40', 'ARL14', 'ARL4C', 'ARL6IP1', 'ARNTL2', 'ARX', 'ASAP1', 'ASAP2', 'ASCL2', 'ASF1B', 'ASGR1', 'ASNS', 'ASRGL1', 'ASS1', 'ATF3', 'ATF5', 'ATP11A', 'ATP1A1', 'ATP1B1', 'ATP1B2', 'ATP2A3', 'ATP8A1', 'B4GALT1', 'B4GALT5', 'BACE2', 'BAIAP2L1', 'BAIAP3', 'BAMBI', 'BBC3', 'BCL2L15', 'BCL3', 'BHLHE40', 'BHLHE41', 'BIK', 'BIRC5', 'BLNK', 'BMF', 'BMP1', 'BMP2', 'BMP7', 'BST2', 'BUB1B', 'BUB1', 'C1QA', 'C1QC', 'C1QL1', 'C2CD4A', 'C2CD4B', 'C2', 'CACNA1A', 'CACNA2D1', 'CACNG4', 'CADM2', 'CADPS', 'CALY', 'CAMK2B', 'CAPG', 'CAPN5', 'CAPN6', 'CARTPT', 'CASP9', 'CAV2', 'CBLB', 'CBLN4', 'CBS', 'CCBE1', 'CCDC34', 'CCDC51', 'CCDC69', 'CCNA1', 'CD14', 'CD276', 'CD300A', 'CD68', 'CD82', 'CD9', 'CDC20', 'CDC42EP3', 'CDCA8', 'CDH17', 'CDH3', 'CDK1', 'CDK5R2', 'CDKN1A', 'CDKN2A', 'CDKN2B', 'CDKN3', 'CDR2L', 'CDX2', 'CEBPB', 'CELF3', 'CENPW', 'CERCAM', 'CFI', 'CHAC1', 'CHDH', 'CHGB', 'CHRNA3', 'CITED2', 'CITED4', 'CKAP2', 'CKAP5', 'CKMT2', 'CKS1B', 'CKS2', 'CLCF1', 'CLDN1', 'CLIC4', 'CLMN', 'CLSPN', 'CMPK2', 'CMTM7', 'CNIH2', 'CNKSR2', 'CNN2', 'CNN3', 'CNTN1', 'COL13A1', 'COL16A1', 'COL18A1', 'COL4A2', 'COL5A3', 'COL9A2', 'COPG1', 'COPZ2', 'CORO1C', 'COTL1', 'COX7A1', 'CPLX2', 'CPPED1', 'CPVL', 'CRABP2', 'CRAT', 'CREB3L1', 'CREB5', 'CRIP1', 'CRLF1', 'CRTAC1', 'CRTAP', 'CRYM', 'CSF1R', 'CSGALNACT1', 'CTSC', 'CTSH', 'CTSO', 'CTSZ', 'CWC25', 'CX3CL1', 'CXADR', 'CXCL12', 'CXCL16', 'CXCL1', 'CYB5A', 'CYP26B1', 'CYP27A1', 'CYTH3', 'DAB2IP', 'DACH1', 'DACT1', 'DAPP1', 'DBN1', 'DBNDD1', 'DCUN1D2', 'DDC', 'DDIT4', 'DDX51', 'DEDD2', 'DHODH', 'DHRS2', 'DIP2A', 'DIRAS2', 'DNAJC12', 'DNAL1', 'DOCK4', 'DOCK6', 'DOCK8', 'DOCK9', 'DOK5', 'DPP4', 'DPP6', 'DSC2', 'DSEL', 'DUSP1', 'DUSP23', 'DUSP26', 'DUSP5', 'ECEL1', 'ECM1', 'ECT2', 'EDN2', 'EEF1D', 'EEF2K', 'EFEMP2', 'EFHD2', 'EFNA1', 'EGFR', 'EGR1', 'EGR4', 'EHD2', 'EHD4', 'EHF', 'ELAVL4', 'EMILIN1', 'EMP3', 'ENAH', 'ENO2', 'ENPP2', 'EPHB2', 'EPHB4', 'EPS8', 'ERBB3', 'ERO1L', 'ERRFI1', 'ETS2', 'EVA1A', 'EVA1B', 'F2RL2', 'F2RL3', 'F2R', 'F5', 'FA2H', 'FAM105A', 'FAM107B', 'FAM129B', 'FAM134B', 'FAM171A1', 'FAM227A', 'FAM43A', 'FAM46A', 'FAM46C', 'FAM84A', 'FBLIM1', 'FBP1', 'FBXL20', 'FBXO2', 'FBXO32', 'FBXO5', 'FBXO6', 'FCER1G', 'FCGRT', 'FERMT1', 'FEV', 'FFAR4', 'FGB', 'FGF13', 'FGFR2', 'FGFR3', 'FHL2', 'FHOD1', 'FJX1', 'FKBP10', 'FKBP11', 'FKBP5', 'FLNA', 'FMNL3', 'FNDC4', 'FNIP2', 'FOSB', 'FOSL2', 'FOXA3', 'FOXC1', 'FOXM1', 'FOXQ1', 'FRZB', 'FSCN1', 'FSTL3', 'FTH1', 'FURIN', 'FUT8', 'FXYD3', 'FXYD5', 'FXYD6', 'G0S2', 'G6PC', 'GAD2', 'GADD45A', 'GADD45B', 'GALE', 'GALM', 'GALNT2', 'GAL', 'GAS6', 'GAS7', 'GATA2', 'GATA4', 'GATSL3', 'GCGR', 'GCH1', 'GCK', 'GCLC', 'GCNT1', 'GCNT3', 'GC', 'GDA', 'GEM', 'GFOD2', 'GFPT2', 'GFRA1', 'GJA1', 'GJB1', 'GK', 'GLIPR1', 'GLRX', 'GLS', 'GMNN', 'GNPAT', 'GOLM1', 'GPC1', 'GPC6', 'GPD1', 'GPR160', 'GPR161', 'GPSM1', 'GPX3', 'GPX8', 'GRASP', 'GREB1', 'GSN', 'GSTM5', 'GSTP1', 'GTF3C3', 'GTPBP10', 'GTSE1', 'GULP1', 'H19', 'HABP2', 'HAND2', 'HDAC9', 'HEPACAM2', 'HEPH', 'HERPUD1', 'HES1', 'HEYL', 'HHEX', 'HIF1A', 'HIF3A', 'HILPDA', 'HIST1H1C', 'HIST1H2BK', 'HIST2H2BE', 'HK1', 'HK2', 'HKDC1', 'HMGB2', 'HMGB3', 'HMGCR', 'HMGCS1', 'HMMR', 'HN1', 'HNF1B', 'HOGA1', 'HOMER3', 'HOPX', 'HOXB2', 'HPCAL1', 'HPGD', 'HPN', 'HS3ST1', 'HSD11B2', 'HSD17B11', 'HSP90B1', 'HSPB1', 'HSPB8', 'HSPH1', 'ID1', 'ID4', 'IDH1', 'IDH2', 'IER2', 'IER3', 'IFIT1', 'IFITM2', 'IFNLR1', 'IGSF10', 'IGSF1', 'IGSF3', 'IL11', 'IL18', 'IL1R2', 'IMPA2', 'INPP5F', 'INSIG1', 'INSM1', 'IP6K1', 'IPCEF1', 'IRAK4', 'IRF1', 'IRF8', 'IRS2', 'IRX1', 'IRX2', 'IRX4', 'ISL1', 'ITGA11', 'ITGA3', 'ITGA9', 'ITGAV', 'ITGB4', 'ITGB6', 'ITGB8', 'ITIH5', 'ITPRIPL2', 'JAG1', 'JARID2', 'JRK', 'JSRP1', 'JUN', 'KANK1', 'KANK2', 'KCNA5', 'KCNAB1', 'KCNE3', 'KCNG1', 'KCNH6', 'KCNJ11', 'KCNJ16', 'KCNJ5', 'KCNJ6', 'KCNK16', 'KCNK3', 'KCNK5', 'KCNMA1', 'KCNN3', 'KCNQ1OT1', 'KCNQ1', 'KCTD12', 'KCTD8', 'KDELC2', 'KDELR3', 'KDM5D', 'KIF11', 'KIF12', 'KIF14', 'KIF21B', 'KIF2C', 'KIF5C', 'KISS1R', 'KLF2', 'KLF4', 'KLF5', 'KLF6', 'KLHDC8A', 'KLHL21', 'KLK11', 'KNSTRN', 'KPNA2', 'KRT17', 'KRTCAP3', 'L2HGDH', 'LAD1', 'LAMB1', 'LAMB2', 'LAPTM5', 'LBH', 'LDLR', 'LECT1', 'LEFTY1', 'LGALS3', 'LGALS9', 'LGI2', 'LHFP', 'LIMA1', 'LIMCH1', 'LIMK2', 'LIMS1', 'LIPH', 'LMO7', 'LONRF2', 'LOXL2', 'LPAR6', 'LRP12', 'LRP1', 'LRRC10B', 'LRRC59', 'LSAMP', 'LTBP4', 'LURAP1L', 'LUZP1', 'LXN', 'LY6E', 'LY6H', 'LYN', 'LYPD1', 'LYPD6B', 'MACC1', 'MAFA', 'MAFB', 'MAFF', 'MAL2', 'MALL', 'MAN1C1', 'MANF', 'MAOB', 'MAP1B', 'MAP2', 'MAP7D2', 'MAPT', 'MARCKSL1', 'MATN2', 'MDFI', 'MDK', 'MECOM', 'MEIS2', 'MERTK', 'METRN', 'METTL20', 'METTL22', 'MFI2', 'MGAT4A', 'MGAT4B', 'MGLL', 'MICAL2', 'MID1IP1', 'MIR143HG', 'MLLT11', 'MLXIPL', 'MMP14', 'MNX1', 'MPZL2', 'MRC1', 'MRC2', 'MSMO1', 'MSN', 'MSRB1', 'MSRB3', 'MTUS1', 'MUM1L1', 'MYC', 'MYH10', 'MYH9', 'MYL12A', 'MYO10', 'MYO1E', 'MYO5B', 'MYRF', 'MYZAP', 'NAA16', 'NCAM1', 'NCAPG', 'NCAPH', 'NCMAP', 'NDRG1', 'NDRG2', 'NDRG4', 'NDUFA4L2', 'NDUFA4', 'NEK8', 'NEURL1B', 'NEURL3', 'NEUROD1', 'NFATC4', 'NFKBIA', 'NFKBIE', 'NINJ1', 'NKD1', 'NKX2-2', 'NLN', 'NMB', 'NMNAT1', 'NOB1', 'NOTCH1', 'NOTCH2', 'NOTCH3', 'NOTCH4', 'NOX4', 'NPHS1', 'NPNT', 'NPTX2', 'NQO1', 'NR0B1', 'NR0B2', 'NR1D1', 'NR2F2', 'NR4A2', 'NR5A2', 'NRP2', 'NTN4', 'NUAK2', 'NUF2', 'NUSAP1', 'OCLN', 'OGDHL', 'OLFML2B', 'OPA3', 'ORM1', 'P4HA3', 'PACSIN2', 'PAH', 'PAK3', 'PALLD', 'PAMR1', 'PAPSS2', 'PARM1', 'PAX6', 'PBK', 'PCDH17', 'PCDH18', 'PCDH9', 'PCK1', 'PCNA', 'PCP4', 'PCSK2', 'PCSK5', 'PDE3A', 'PDE3B', 'PDE8B', 'PDGFA', 'PDGFD', 'PDIA4', 'PDLIM1', 'PDLIM4', 'PDLIM7', 'PDP2', 'PDX1', 'PEMT', 'PER3', 'PEX13', 'PFKFB3', 'PGBD5', 'PGM1', 'PGM2L1', 'PHF19', 'PHGDH', 'PHGR1', 'PHLDA2', 'PHLDB1', 'PID1', 'PIEZO1', 'PIK3AP1', 'PIK3R1', 'PIM1', 'PKIB', 'PKNOX2', 'PKP2', 'PLA2G4C', 'PLA2G7', 'PLAA', 'PLAGL1', 'PLAT', 'PLCXD3', 'PLEKHA2', 'PLEKHB1', 'PLEKHG2', 'PLEKHO1', 'PLIN5', 'PLK2', 'PLLP', 'PLOD2', 'PLSCR1', 'PLXDC1', 'PLXNA1', 'PLXNA2', 'PLXND1', 'PMAIP1', 'PNMA2', 'PNRC1', 'POC1A', 'POLD4', 'PON2', 'POU2F2', 'PPARGC1A', 'PPIC', 'PPIF', 'PPL', 'PPP1R15A', 'PPP1R18', 'PPP1R1A', 'PPP1R1B', 'PPP2R2B', 'PRC1', 'PRELP', 'PRKCDBP', 'PRKG1', 'PRKX', 'PRLHR', 'PRMT6', 'PRR11', 'PRR15L', 'PRSS22', 'PRSS23', 'PRSS8', 'PTAFR', 'PTGDS', 'PTGES', 'PTGFR', 'PTK7', 'PTPN14', 'PTPN3', 'PTPRN', 'PTPRT', 'PTRF', 'PTTG1', 'PVRL3', 'PVRL4', 'PYCARD', 'PYCR1', 'PYROXD2', 'QPRT', 'QSOX1', 'RAB11FIP1', 'RAB33A', 'RAB3B', 'RAB42', 'RACGAP1', 'RAMP1', 'RAP1GAP', 'RAPGEF5', 'RARG', 'RARRES1', 'RASA3', 'RASEF', 'RASGRP3', 'RASL12', 'RASSF4', 'RASSF9', 'RBM38', 'RBP1', 'RBPMS', 'RCL1', 'RCN3', 'RDH10', 'RDX', 'RELB', 'RETSAT', 'RFTN1', 'RFX6', 'RGS5', 'RHBDF1', 'RHOB', 'RHOC', 'RHOD', 'RHOV', 'RHPN2', 'RINL', 'RNASE6', 'RNF114', 'RNF157', 'RNF24', 'RNF32', 'ROBO1', 'ROR2', 'RPL22L1', 'RPS6KA2', 'RRAGD', 'RSU1', 'S100A10', 'S100A11', 'S100A16', 'S1PR3', 'SAMHD1', 'SCG2', 'SCG3', 'SCN1B', 'SCNN1A', 'SCTR', 'SDC2', 'SDS', 'SEL1L3', 'SEMA3B', 'SEMA3C', 'SEMA4B', 'SEMA6A', 'SEMA7A', 'SEPT3', 'SEPT6', 'SEPT9', 'SERPINA6', 'SERPINE2', 'SERPINF1', 'SERPINI1', 'SERTM1', 'SETX', 'SEZ6L', 'SFMBT2', 'SFN', 'SFRP1', 'SFRP5', 'SGOL1', 'SGSM1', 'SGSM3', 'SH3BGRL3', 'SH3BP4', 'SH3KBP1', 'SH3PXD2A', 'SHANK2', 'SHC1', 'SHROOM1', 'SHROOM3', 'SIRPA', 'SIX4', 'SKIL', 'SLC12A2', 'SLC12A7', 'SLC15A4', 'SLC16A3', 'SLC16A5', 'SLC16A7', 'SLC16A9', 'SLC1A5', 'SLC22A17', 'SLC22A3', 'SLC25A43', 'SLC26A4', 'SLC29A4', 'SLC2A13', 'SLC30A8', 'SLC38A1', 'SLC38A2', 'SLC38A5', 'SLC39A14', 'SLC39A5', 'SLC40A1', 'SLC43A3', 'SLC44A2', 'SLC45A3', 'SLC4A4', 'SLC4A5', 'SL
#> ℹ [2025-07-26 07:18:24] Data 1/5 of the srt_list has been log-normalized.
#> ℹ [2025-07-26 07:18:24] Perform FindVariableFeatures on the data 1/5 of the srt_list...
#> Warning: The following features were labelled as variable in 'var.features' but had no corresponding rank in `var.features.rank` and will therefore be ignored: 'ABCC3', 'ABCC8', 'ABCC9', 'ABCG1', 'ABCG2', 'ABHD2', 'ABHD4', 'ABI3', 'ACHE', 'ACP5', 'ACSL4', 'ACSS1', 'ACTN1', 'ACTN4', 'ADAM8', 'ADAM9', 'ADAMTS5', 'ADAMTS9', 'ADAMTSL2', 'ADAP2', 'ADCY1', 'ADCY3', 'ADCY5', 'ADM2', 'ADRA2A', 'ADRBK2', 'AEN', 'AGR2', 'AGRN', 'AGTRAP', 'AHNAK', 'AHR', 'AKAP7', 'ALDH1A1', 'ALDH1A2', 'ALDH1B1', 'ALOX5AP', 'ALS2CL', 'AMOTL2', 'AMT', 'ANK1', 'ANKRD1', 'ANKRD37', 'ANLN', 'ANO6', 'ANXA13', 'ANXA3', 'ANXA9', 'AP5Z1', 'AQP3', 'ARC', 'ARG2', 'ARHGAP22', 'ARHGAP23', 'ARHGAP29', 'ARHGEF10L', 'ARHGEF2', 'ARHGEF39', 'ARHGEF40', 'ARL14', 'ARL4C', 'ARL6IP1', 'ARNTL2', 'ARX', 'ASAP1', 'ASAP2', 'ASCL2', 'ASF1B', 'ASGR1', 'ASNS', 'ASRGL1', 'ASS1', 'ATF3', 'ATF5', 'ATP11A', 'ATP1A1', 'ATP1B1', 'ATP1B2', 'ATP2A3', 'ATP8A1', 'B4GALT1', 'B4GALT5', 'BACE2', 'BAIAP2L1', 'BAIAP3', 'BAMBI', 'BBC3', 'BCL2L15', 'BCL3', 'BHLHE40', 'BHLHE41', 'BIK', 'BIRC5', 'BLNK', 'BMF', 'BMP1', 'BMP2', 'BMP7', 'BST2', 'BUB1B', 'BUB1', 'C1QA', 'C1QC', 'C1QL1', 'C2CD4A', 'C2CD4B', 'C2', 'CACNA1A', 'CACNA2D1', 'CACNG4', 'CADM2', 'CADPS', 'CALY', 'CAMK2B', 'CAPG', 'CAPN5', 'CAPN6', 'CARTPT', 'CASP9', 'CAV2', 'CBLB', 'CBLN4', 'CBS', 'CCBE1', 'CCDC34', 'CCDC51', 'CCDC69', 'CCNA1', 'CD14', 'CD276', 'CD300A', 'CD68', 'CD82', 'CD9', 'CDC20', 'CDC42EP3', 'CDCA8', 'CDH17', 'CDH3', 'CDK1', 'CDK5R2', 'CDKN1A', 'CDKN2A', 'CDKN2B', 'CDKN3', 'CDR2L', 'CDX2', 'CEBPB', 'CELF3', 'CENPW', 'CERCAM', 'CFI', 'CHAC1', 'CHDH', 'CHGB', 'CHRNA3', 'CITED2', 'CITED4', 'CKAP2', 'CKAP5', 'CKMT2', 'CKS1B', 'CKS2', 'CLCF1', 'CLDN1', 'CLIC4', 'CLMN', 'CLSPN', 'CMPK2', 'CMTM7', 'CNIH2', 'CNKSR2', 'CNN2', 'CNN3', 'CNTN1', 'COL13A1', 'COL16A1', 'COL18A1', 'COL4A2', 'COL5A3', 'COL9A2', 'COPG1', 'COPZ2', 'CORO1C', 'COTL1', 'COX7A1', 'CPLX2', 'CPPED1', 'CPVL', 'CRABP2', 'CRAT', 'CREB3L1', 'CREB5', 'CRIP1', 'CRLF1', 'CRTAC1', 'CRTAP', 'CRYM', 'CSF1R', 'CSGALNACT1', 'CTSC', 'CTSH', 'CTSO', 'CTSZ', 'CWC25', 'CX3CL1', 'CXADR', 'CXCL12', 'CXCL16', 'CXCL1', 'CYB5A', 'CYP26B1', 'CYP27A1', 'CYTH3', 'DAB2IP', 'DACH1', 'DACT1', 'DAPP1', 'DBN1', 'DBNDD1', 'DCUN1D2', 'DDC', 'DDIT4', 'DDX51', 'DEDD2', 'DHODH', 'DHRS2', 'DIP2A', 'DIRAS2', 'DNAJC12', 'DNAL1', 'DOCK4', 'DOCK6', 'DOCK8', 'DOCK9', 'DOK5', 'DPP4', 'DPP6', 'DSC2', 'DSEL', 'DUSP1', 'DUSP23', 'DUSP26', 'DUSP5', 'ECEL1', 'ECM1', 'ECT2', 'EDN2', 'EEF1D', 'EEF2K', 'EFEMP2', 'EFHD2', 'EFNA1', 'EGFR', 'EGR1', 'EGR4', 'EHD2', 'EHD4', 'EHF', 'ELAVL4', 'EMILIN1', 'EMP3', 'ENAH', 'ENO2', 'ENPP2', 'EPHB2', 'EPHB4', 'EPS8', 'ERBB3', 'ERO1L', 'ERRFI1', 'ETS2', 'EVA1A', 'EVA1B', 'F2RL2', 'F2RL3', 'F2R', 'F5', 'FA2H', 'FAM105A', 'FAM107B', 'FAM129B', 'FAM134B', 'FAM171A1', 'FAM227A', 'FAM43A', 'FAM46A', 'FAM46C', 'FAM84A', 'FBLIM1', 'FBP1', 'FBXL20', 'FBXO2', 'FBXO32', 'FBXO5', 'FBXO6', 'FCER1G', 'FCGRT', 'FERMT1', 'FEV', 'FFAR4', 'FGB', 'FGF13', 'FGFR2', 'FGFR3', 'FHL2', 'FHOD1', 'FJX1', 'FKBP10', 'FKBP11', 'FKBP5', 'FLNA', 'FMNL3', 'FNDC4', 'FNIP2', 'FOSB', 'FOSL2', 'FOXA3', 'FOXC1', 'FOXM1', 'FOXQ1', 'FRZB', 'FSCN1', 'FSTL3', 'FTH1', 'FURIN', 'FUT8', 'FXYD3', 'FXYD5', 'FXYD6', 'G0S2', 'G6PC', 'GAD2', 'GADD45A', 'GADD45B', 'GALE', 'GALM', 'GALNT2', 'GAL', 'GAS6', 'GAS7', 'GATA2', 'GATA4', 'GATSL3', 'GCGR', 'GCH1', 'GCK', 'GCLC', 'GCNT1', 'GCNT3', 'GC', 'GDA', 'GEM', 'GFOD2', 'GFPT2', 'GFRA1', 'GJA1', 'GJB1', 'GK', 'GLIPR1', 'GLRX', 'GLS', 'GMNN', 'GNPAT', 'GOLM1', 'GPC1', 'GPC6', 'GPD1', 'GPR160', 'GPR161', 'GPSM1', 'GPX3', 'GPX8', 'GRASP', 'GREB1', 'GSN', 'GSTM5', 'GSTP1', 'GTF3C3', 'GTPBP10', 'GTSE1', 'GULP1', 'H19', 'HABP2', 'HAND2', 'HDAC9', 'HEPACAM2', 'HEPH', 'HERPUD1', 'HES1', 'HEYL', 'HHEX', 'HIF1A', 'HIF3A', 'HILPDA', 'HIST1H1C', 'HIST1H2BK', 'HIST2H2BE', 'HK1', 'HK2', 'HKDC1', 'HMGB2', 'HMGB3', 'HMGCR', 'HMGCS1', 'HMMR', 'HN1', 'HNF1B', 'HOGA1', 'HOMER3', 'HOPX', 'HOXB2', 'HPCAL1', 'HPGD', 'HPN', 'HS3ST1', 'HSD11B2', 'HSD17B11', 'HSP90B1', 'HSPB1', 'HSPB8', 'HSPH1', 'ID1', 'ID4', 'IDH1', 'IDH2', 'IER2', 'IER3', 'IFIT1', 'IFITM2', 'IFNLR1', 'IGSF10', 'IGSF1', 'IGSF3', 'IL11', 'IL18', 'IL1R2', 'IMPA2', 'INPP5F', 'INSIG1', 'INSM1', 'IP6K1', 'IPCEF1', 'IRAK4', 'IRF1', 'IRF8', 'IRS2', 'IRX1', 'IRX2', 'IRX4', 'ISL1', 'ITGA11', 'ITGA3', 'ITGA9', 'ITGAV', 'ITGB4', 'ITGB6', 'ITGB8', 'ITIH5', 'ITPRIPL2', 'JAG1', 'JARID2', 'JRK', 'JSRP1', 'JUN', 'KANK1', 'KANK2', 'KCNA5', 'KCNAB1', 'KCNE3', 'KCNG1', 'KCNH6', 'KCNJ11', 'KCNJ16', 'KCNJ5', 'KCNJ6', 'KCNK16', 'KCNK3', 'KCNK5', 'KCNMA1', 'KCNN3', 'KCNQ1OT1', 'KCNQ1', 'KCTD12', 'KCTD8', 'KDELC2', 'KDELR3', 'KDM5D', 'KIF11', 'KIF12', 'KIF14', 'KIF21B', 'KIF2C', 'KIF5C', 'KISS1R', 'KLF2', 'KLF4', 'KLF5', 'KLF6', 'KLHDC8A', 'KLHL21', 'KLK11', 'KNSTRN', 'KPNA2', 'KRT17', 'KRTCAP3', 'L2HGDH', 'LAD1', 'LAMB1', 'LAMB2', 'LAPTM5', 'LBH', 'LDLR', 'LECT1', 'LEFTY1', 'LGALS3', 'LGALS9', 'LGI2', 'LHFP', 'LIMA1', 'LIMCH1', 'LIMK2', 'LIMS1', 'LIPH', 'LMO7', 'LONRF2', 'LOXL2', 'LPAR6', 'LRP12', 'LRP1', 'LRRC10B', 'LRRC59', 'LSAMP', 'LTBP4', 'LURAP1L', 'LUZP1', 'LXN', 'LY6E', 'LY6H', 'LYN', 'LYPD1', 'LYPD6B', 'MACC1', 'MAFA', 'MAFB', 'MAFF', 'MAL2', 'MALL', 'MAN1C1', 'MANF', 'MAOB', 'MAP1B', 'MAP2', 'MAP7D2', 'MAPT', 'MARCKSL1', 'MATN2', 'MDFI', 'MDK', 'MECOM', 'MEIS2', 'MERTK', 'METRN', 'METTL20', 'METTL22', 'MFI2', 'MGAT4A', 'MGAT4B', 'MGLL', 'MICAL2', 'MID1IP1', 'MIR143HG', 'MLLT11', 'MLXIPL', 'MMP14', 'MNX1', 'MPZL2', 'MRC1', 'MRC2', 'MSMO1', 'MSN', 'MSRB1', 'MSRB3', 'MTUS1', 'MUM1L1', 'MYC', 'MYH10', 'MYH9', 'MYL12A', 'MYO10', 'MYO1E', 'MYO5B', 'MYRF', 'MYZAP', 'NAA16', 'NCAM1', 'NCAPG', 'NCAPH', 'NCMAP', 'NDRG1', 'NDRG2', 'NDRG4', 'NDUFA4L2', 'NDUFA4', 'NEK8', 'NEURL1B', 'NEURL3', 'NEUROD1', 'NFATC4', 'NFKBIA', 'NFKBIE', 'NINJ1', 'NKD1', 'NKX2-2', 'NLN', 'NMB', 'NMNAT1', 'NOB1', 'NOTCH1', 'NOTCH2', 'NOTCH3', 'NOTCH4', 'NOX4', 'NPHS1', 'NPNT', 'NPTX2', 'NQO1', 'NR0B1', 'NR0B2', 'NR1D1', 'NR2F2', 'NR4A2', 'NR5A2', 'NRP2', 'NTN4', 'NUAK2', 'NUF2', 'NUSAP1', 'OCLN', 'OGDHL', 'OLFML2B', 'OPA3', 'ORM1', 'P4HA3', 'PACSIN2', 'PAH', 'PAK3', 'PALLD', 'PAMR1', 'PAPSS2', 'PARM1', 'PAX6', 'PBK', 'PCDH17', 'PCDH18', 'PCDH9', 'PCK1', 'PCNA', 'PCP4', 'PCSK2', 'PCSK5', 'PDE3A', 'PDE3B', 'PDE8B', 'PDGFA', 'PDGFD', 'PDIA4', 'PDLIM1', 'PDLIM4', 'PDLIM7', 'PDP2', 'PDX1', 'PEMT', 'PER3', 'PEX13', 'PFKFB3', 'PGBD5', 'PGM1', 'PGM2L1', 'PHF19', 'PHGDH', 'PHGR1', 'PHLDA2', 'PHLDB1', 'PID1', 'PIEZO1', 'PIK3AP1', 'PIK3R1', 'PIM1', 'PKIB', 'PKNOX2', 'PKP2', 'PLA2G4C', 'PLA2G7', 'PLAA', 'PLAGL1', 'PLAT', 'PLCXD3', 'PLEKHA2', 'PLEKHB1', 'PLEKHG2', 'PLEKHO1', 'PLIN5', 'PLK2', 'PLLP', 'PLOD2', 'PLSCR1', 'PLXDC1', 'PLXNA1', 'PLXNA2', 'PLXND1', 'PMAIP1', 'PNMA2', 'PNRC1', 'POC1A', 'POLD4', 'PON2', 'POU2F2', 'PPARGC1A', 'PPIC', 'PPIF', 'PPL', 'PPP1R15A', 'PPP1R18', 'PPP1R1A', 'PPP1R1B', 'PPP2R2B', 'PRC1', 'PRELP', 'PRKCDBP', 'PRKG1', 'PRKX', 'PRLHR', 'PRMT6', 'PRR11', 'PRR15L', 'PRSS22', 'PRSS23', 'PRSS8', 'PTAFR', 'PTGDS', 'PTGES', 'PTGFR', 'PTK7', 'PTPN14', 'PTPN3', 'PTPRN', 'PTPRT', 'PTRF', 'PTTG1', 'PVRL3', 'PVRL4', 'PYCARD', 'PYCR1', 'PYROXD2', 'QPRT', 'QSOX1', 'RAB11FIP1', 'RAB33A', 'RAB3B', 'RAB42', 'RACGAP1', 'RAMP1', 'RAP1GAP', 'RAPGEF5', 'RARG', 'RARRES1', 'RASA3', 'RASEF', 'RASGRP3', 'RASL12', 'RASSF4', 'RASSF9', 'RBM38', 'RBP1', 'RBPMS', 'RCL1', 'RCN3', 'RDH10', 'RDX', 'RELB', 'RETSAT', 'RFTN1', 'RFX6', 'RGS5', 'RHBDF1', 'RHOB', 'RHOC', 'RHOD', 'RHOV', 'RHPN2', 'RINL', 'RNASE6', 'RNF114', 'RNF157', 'RNF24', 'RNF32', 'ROBO1', 'ROR2', 'RPL22L1', 'RPS6KA2', 'RRAGD', 'RSU1', 'S100A10', 'S100A11', 'S100A16', 'S1PR3', 'SAMHD1', 'SCG2', 'SCG3', 'SCN1B', 'SCNN1A', 'SCTR', 'SDC2', 'SDS', 'SEL1L3', 'SEMA3B', 'SEMA3C', 'SEMA4B', 'SEMA6A', 'SEMA7A', 'SEPT3', 'SEPT6', 'SEPT9', 'SERPINA6', 'SERPINE2', 'SERPINF1', 'SERPINI1', 'SERTM1', 'SETX', 'SEZ6L', 'SFMBT2', 'SFN', 'SFRP1', 'SFRP5', 'SGOL1', 'SGSM1', 'SGSM3', 'SH3BGRL3', 'SH3BP4', 'SH3KBP1', 'SH3PXD2A', 'SHANK2', 'SHC1', 'SHROOM1', 'SHROOM3', 'SIRPA', 'SIX4', 'SKIL', 'SLC12A2', 'SLC12A7', 'SLC15A4', 'SLC16A3', 'SLC16A5', 'SLC16A7', 'SLC16A9', 'SLC1A5', 'SLC22A17', 'SLC22A3', 'SLC25A43', 'SLC26A4', 'SLC29A4', 'SLC2A13', 'SLC30A8', 'SLC38A1', 'SLC38A2', 'SLC38A5', 'SLC39A14', 'SLC39A5', 'SLC40A1', 'SLC43A3', 'SLC44A2', 'SLC45A3', 'SLC4A4', 'SLC4A5', 'SL
#> ℹ [2025-07-26 07:18:25] Data 2/5 of the srt_list has been log-normalized.
#> ℹ [2025-07-26 07:18:25] Perform FindVariableFeatures on the data 2/5 of the srt_list...
#> Warning: The following features were labelled as variable in 'var.features' but had no corresponding rank in `var.features.rank` and will therefore be ignored: 'ABCC3', 'ABCC8', 'ABCC9', 'ABCG1', 'ABCG2', 'ABHD2', 'ABHD4', 'ABI3', 'ACHE', 'ACP5', 'ACSL4', 'ACSS1', 'ACTN1', 'ACTN4', 'ADAM8', 'ADAM9', 'ADAMTS5', 'ADAMTS9', 'ADAMTSL2', 'ADAP2', 'ADCY1', 'ADCY3', 'ADCY5', 'ADM2', 'ADRA2A', 'ADRBK2', 'AEN', 'AGR2', 'AGRN', 'AGTRAP', 'AHNAK', 'AHR', 'AKAP7', 'ALDH1A1', 'ALDH1A2', 'ALDH1B1', 'ALOX5AP', 'ALS2CL', 'AMOTL2', 'AMT', 'ANK1', 'ANKRD1', 'ANKRD37', 'ANLN', 'ANO6', 'ANXA13', 'ANXA3', 'ANXA9', 'AP5Z1', 'AQP3', 'ARC', 'ARG2', 'ARHGAP22', 'ARHGAP23', 'ARHGAP29', 'ARHGEF10L', 'ARHGEF2', 'ARHGEF39', 'ARHGEF40', 'ARL14', 'ARL4C', 'ARL6IP1', 'ARNTL2', 'ARX', 'ASAP1', 'ASAP2', 'ASCL2', 'ASF1B', 'ASGR1', 'ASNS', 'ASRGL1', 'ASS1', 'ATF3', 'ATF5', 'ATP11A', 'ATP1A1', 'ATP1B1', 'ATP1B2', 'ATP2A3', 'ATP8A1', 'B4GALT1', 'B4GALT5', 'BACE2', 'BAIAP2L1', 'BAIAP3', 'BAMBI', 'BBC3', 'BCL2L15', 'BCL3', 'BHLHE40', 'BHLHE41', 'BIK', 'BIRC5', 'BLNK', 'BMF', 'BMP1', 'BMP2', 'BMP7', 'BST2', 'BUB1B', 'BUB1', 'C1QA', 'C1QC', 'C1QL1', 'C2CD4A', 'C2CD4B', 'C2', 'CACNA1A', 'CACNA2D1', 'CACNG4', 'CADM2', 'CADPS', 'CALY', 'CAMK2B', 'CAPG', 'CAPN5', 'CAPN6', 'CARTPT', 'CASP9', 'CAV2', 'CBLB', 'CBLN4', 'CBS', 'CCBE1', 'CCDC34', 'CCDC51', 'CCDC69', 'CCNA1', 'CD14', 'CD276', 'CD300A', 'CD68', 'CD82', 'CD9', 'CDC20', 'CDC42EP3', 'CDCA8', 'CDH17', 'CDH3', 'CDK1', 'CDK5R2', 'CDKN1A', 'CDKN2A', 'CDKN2B', 'CDKN3', 'CDR2L', 'CDX2', 'CEBPB', 'CELF3', 'CENPW', 'CERCAM', 'CFI', 'CHAC1', 'CHDH', 'CHGB', 'CHRNA3', 'CITED2', 'CITED4', 'CKAP2', 'CKAP5', 'CKMT2', 'CKS1B', 'CKS2', 'CLCF1', 'CLDN1', 'CLIC4', 'CLMN', 'CLSPN', 'CMPK2', 'CMTM7', 'CNIH2', 'CNKSR2', 'CNN2', 'CNN3', 'CNTN1', 'COL13A1', 'COL16A1', 'COL18A1', 'COL4A2', 'COL5A3', 'COL9A2', 'COPG1', 'COPZ2', 'CORO1C', 'COTL1', 'COX7A1', 'CPLX2', 'CPPED1', 'CPVL', 'CRABP2', 'CRAT', 'CREB3L1', 'CREB5', 'CRIP1', 'CRLF1', 'CRTAC1', 'CRTAP', 'CRYM', 'CSF1R', 'CSGALNACT1', 'CTSC', 'CTSH', 'CTSO', 'CTSZ', 'CWC25', 'CX3CL1', 'CXADR', 'CXCL12', 'CXCL16', 'CXCL1', 'CYB5A', 'CYP26B1', 'CYP27A1', 'CYTH3', 'DAB2IP', 'DACH1', 'DACT1', 'DAPP1', 'DBN1', 'DBNDD1', 'DCUN1D2', 'DDC', 'DDIT4', 'DDX51', 'DEDD2', 'DHODH', 'DHRS2', 'DIP2A', 'DIRAS2', 'DNAJC12', 'DNAL1', 'DOCK4', 'DOCK6', 'DOCK8', 'DOCK9', 'DOK5', 'DPP4', 'DPP6', 'DSC2', 'DSEL', 'DUSP1', 'DUSP23', 'DUSP26', 'DUSP5', 'ECEL1', 'ECM1', 'ECT2', 'EDN2', 'EEF1D', 'EEF2K', 'EFEMP2', 'EFHD2', 'EFNA1', 'EGFR', 'EGR1', 'EGR4', 'EHD2', 'EHD4', 'EHF', 'ELAVL4', 'EMILIN1', 'EMP3', 'ENAH', 'ENO2', 'ENPP2', 'EPHB2', 'EPHB4', 'EPS8', 'ERBB3', 'ERO1L', 'ERRFI1', 'ETS2', 'EVA1A', 'EVA1B', 'F2RL2', 'F2RL3', 'F2R', 'F5', 'FA2H', 'FAM105A', 'FAM107B', 'FAM129B', 'FAM134B', 'FAM171A1', 'FAM227A', 'FAM43A', 'FAM46A', 'FAM46C', 'FAM84A', 'FBLIM1', 'FBP1', 'FBXL20', 'FBXO2', 'FBXO32', 'FBXO5', 'FBXO6', 'FCER1G', 'FCGRT', 'FERMT1', 'FEV', 'FFAR4', 'FGB', 'FGF13', 'FGFR2', 'FGFR3', 'FHL2', 'FHOD1', 'FJX1', 'FKBP10', 'FKBP11', 'FKBP5', 'FLNA', 'FMNL3', 'FNDC4', 'FNIP2', 'FOSB', 'FOSL2', 'FOXA3', 'FOXC1', 'FOXM1', 'FOXQ1', 'FRZB', 'FSCN1', 'FSTL3', 'FTH1', 'FURIN', 'FUT8', 'FXYD3', 'FXYD5', 'FXYD6', 'G0S2', 'G6PC', 'GAD2', 'GADD45A', 'GADD45B', 'GALE', 'GALM', 'GALNT2', 'GAL', 'GAS6', 'GAS7', 'GATA2', 'GATA4', 'GATSL3', 'GCGR', 'GCH1', 'GCK', 'GCLC', 'GCNT1', 'GCNT3', 'GC', 'GDA', 'GEM', 'GFOD2', 'GFPT2', 'GFRA1', 'GJA1', 'GJB1', 'GK', 'GLIPR1', 'GLRX', 'GLS', 'GMNN', 'GNPAT', 'GOLM1', 'GPC1', 'GPC6', 'GPD1', 'GPR160', 'GPR161', 'GPSM1', 'GPX3', 'GPX8', 'GRASP', 'GREB1', 'GSN', 'GSTM5', 'GSTP1', 'GTF3C3', 'GTPBP10', 'GTSE1', 'GULP1', 'H19', 'HABP2', 'HAND2', 'HDAC9', 'HEPACAM2', 'HEPH', 'HERPUD1', 'HES1', 'HEYL', 'HHEX', 'HIF1A', 'HIF3A', 'HILPDA', 'HIST1H1C', 'HIST1H2BK', 'HIST2H2BE', 'HK1', 'HK2', 'HKDC1', 'HMGB2', 'HMGB3', 'HMGCR', 'HMGCS1', 'HMMR', 'HN1', 'HNF1B', 'HOGA1', 'HOMER3', 'HOPX', 'HOXB2', 'HPCAL1', 'HPGD', 'HPN', 'HS3ST1', 'HSD11B2', 'HSD17B11', 'HSP90B1', 'HSPB1', 'HSPB8', 'HSPH1', 'ID1', 'ID4', 'IDH1', 'IDH2', 'IER2', 'IER3', 'IFIT1', 'IFITM2', 'IFNLR1', 'IGSF10', 'IGSF1', 'IGSF3', 'IL11', 'IL18', 'IL1R2', 'IMPA2', 'INPP5F', 'INSIG1', 'INSM1', 'IP6K1', 'IPCEF1', 'IRAK4', 'IRF1', 'IRF8', 'IRS2', 'IRX1', 'IRX2', 'IRX4', 'ISL1', 'ITGA11', 'ITGA3', 'ITGA9', 'ITGAV', 'ITGB4', 'ITGB6', 'ITGB8', 'ITIH5', 'ITPRIPL2', 'JAG1', 'JARID2', 'JRK', 'JSRP1', 'JUN', 'KANK1', 'KANK2', 'KCNA5', 'KCNAB1', 'KCNE3', 'KCNG1', 'KCNH6', 'KCNJ11', 'KCNJ16', 'KCNJ5', 'KCNJ6', 'KCNK16', 'KCNK3', 'KCNK5', 'KCNMA1', 'KCNN3', 'KCNQ1OT1', 'KCNQ1', 'KCTD12', 'KCTD8', 'KDELC2', 'KDELR3', 'KDM5D', 'KIF11', 'KIF12', 'KIF14', 'KIF21B', 'KIF2C', 'KIF5C', 'KISS1R', 'KLF2', 'KLF4', 'KLF5', 'KLF6', 'KLHDC8A', 'KLHL21', 'KLK11', 'KNSTRN', 'KPNA2', 'KRT17', 'KRTCAP3', 'L2HGDH', 'LAD1', 'LAMB1', 'LAMB2', 'LAPTM5', 'LBH', 'LDLR', 'LECT1', 'LEFTY1', 'LGALS3', 'LGALS9', 'LGI2', 'LHFP', 'LIMA1', 'LIMCH1', 'LIMK2', 'LIMS1', 'LIPH', 'LMO7', 'LONRF2', 'LOXL2', 'LPAR6', 'LRP12', 'LRP1', 'LRRC10B', 'LRRC59', 'LSAMP', 'LTBP4', 'LURAP1L', 'LUZP1', 'LXN', 'LY6E', 'LY6H', 'LYN', 'LYPD1', 'LYPD6B', 'MACC1', 'MAFA', 'MAFB', 'MAFF', 'MAL2', 'MALL', 'MAN1C1', 'MANF', 'MAOB', 'MAP1B', 'MAP2', 'MAP7D2', 'MAPT', 'MARCKSL1', 'MATN2', 'MDFI', 'MDK', 'MECOM', 'MEIS2', 'MERTK', 'METRN', 'METTL20', 'METTL22', 'MFI2', 'MGAT4A', 'MGAT4B', 'MGLL', 'MICAL2', 'MID1IP1', 'MIR143HG', 'MLLT11', 'MLXIPL', 'MMP14', 'MNX1', 'MPZL2', 'MRC1', 'MRC2', 'MSMO1', 'MSN', 'MSRB1', 'MSRB3', 'MTUS1', 'MUM1L1', 'MYC', 'MYH10', 'MYH9', 'MYL12A', 'MYO10', 'MYO1E', 'MYO5B', 'MYRF', 'MYZAP', 'NAA16', 'NCAM1', 'NCAPG', 'NCAPH', 'NCMAP', 'NDRG1', 'NDRG2', 'NDRG4', 'NDUFA4L2', 'NDUFA4', 'NEK8', 'NEURL1B', 'NEURL3', 'NEUROD1', 'NFATC4', 'NFKBIA', 'NFKBIE', 'NINJ1', 'NKD1', 'NKX2-2', 'NLN', 'NMB', 'NMNAT1', 'NOB1', 'NOTCH1', 'NOTCH2', 'NOTCH3', 'NOTCH4', 'NOX4', 'NPHS1', 'NPNT', 'NPTX2', 'NQO1', 'NR0B1', 'NR0B2', 'NR1D1', 'NR2F2', 'NR4A2', 'NR5A2', 'NRP2', 'NTN4', 'NUAK2', 'NUF2', 'NUSAP1', 'OCLN', 'OGDHL', 'OLFML2B', 'OPA3', 'ORM1', 'P4HA3', 'PACSIN2', 'PAH', 'PAK3', 'PALLD', 'PAMR1', 'PAPSS2', 'PARM1', 'PAX6', 'PBK', 'PCDH17', 'PCDH18', 'PCDH9', 'PCK1', 'PCNA', 'PCP4', 'PCSK2', 'PCSK5', 'PDE3A', 'PDE3B', 'PDE8B', 'PDGFA', 'PDGFD', 'PDIA4', 'PDLIM1', 'PDLIM4', 'PDLIM7', 'PDP2', 'PDX1', 'PEMT', 'PER3', 'PEX13', 'PFKFB3', 'PGBD5', 'PGM1', 'PGM2L1', 'PHF19', 'PHGDH', 'PHGR1', 'PHLDA2', 'PHLDB1', 'PID1', 'PIEZO1', 'PIK3AP1', 'PIK3R1', 'PIM1', 'PKIB', 'PKNOX2', 'PKP2', 'PLA2G4C', 'PLA2G7', 'PLAA', 'PLAGL1', 'PLAT', 'PLCXD3', 'PLEKHA2', 'PLEKHB1', 'PLEKHG2', 'PLEKHO1', 'PLIN5', 'PLK2', 'PLLP', 'PLOD2', 'PLSCR1', 'PLXDC1', 'PLXNA1', 'PLXNA2', 'PLXND1', 'PMAIP1', 'PNMA2', 'PNRC1', 'POC1A', 'POLD4', 'PON2', 'POU2F2', 'PPARGC1A', 'PPIC', 'PPIF', 'PPL', 'PPP1R15A', 'PPP1R18', 'PPP1R1A', 'PPP1R1B', 'PPP2R2B', 'PRC1', 'PRELP', 'PRKCDBP', 'PRKG1', 'PRKX', 'PRLHR', 'PRMT6', 'PRR11', 'PRR15L', 'PRSS22', 'PRSS23', 'PRSS8', 'PTAFR', 'PTGDS', 'PTGES', 'PTGFR', 'PTK7', 'PTPN14', 'PTPN3', 'PTPRN', 'PTPRT', 'PTRF', 'PTTG1', 'PVRL3', 'PVRL4', 'PYCARD', 'PYCR1', 'PYROXD2', 'QPRT', 'QSOX1', 'RAB11FIP1', 'RAB33A', 'RAB3B', 'RAB42', 'RACGAP1', 'RAMP1', 'RAP1GAP', 'RAPGEF5', 'RARG', 'RARRES1', 'RASA3', 'RASEF', 'RASGRP3', 'RASL12', 'RASSF4', 'RASSF9', 'RBM38', 'RBP1', 'RBPMS', 'RCL1', 'RCN3', 'RDH10', 'RDX', 'RELB', 'RETSAT', 'RFTN1', 'RFX6', 'RGS5', 'RHBDF1', 'RHOB', 'RHOC', 'RHOD', 'RHOV', 'RHPN2', 'RINL', 'RNASE6', 'RNF114', 'RNF157', 'RNF24', 'RNF32', 'ROBO1', 'ROR2', 'RPL22L1', 'RPS6KA2', 'RRAGD', 'RSU1', 'S100A10', 'S100A11', 'S100A16', 'S1PR3', 'SAMHD1', 'SCG2', 'SCG3', 'SCN1B', 'SCNN1A', 'SCTR', 'SDC2', 'SDS', 'SEL1L3', 'SEMA3B', 'SEMA3C', 'SEMA4B', 'SEMA6A', 'SEMA7A', 'SEPT3', 'SEPT6', 'SEPT9', 'SERPINA6', 'SERPINE2', 'SERPINF1', 'SERPINI1', 'SERTM1', 'SETX', 'SEZ6L', 'SFMBT2', 'SFN', 'SFRP1', 'SFRP5', 'SGOL1', 'SGSM1', 'SGSM3', 'SH3BGRL3', 'SH3BP4', 'SH3KBP1', 'SH3PXD2A', 'SHANK2', 'SHC1', 'SHROOM1', 'SHROOM3', 'SIRPA', 'SIX4', 'SKIL', 'SLC12A2', 'SLC12A7', 'SLC15A4', 'SLC16A3', 'SLC16A5', 'SLC16A7', 'SLC16A9', 'SLC1A5', 'SLC22A17', 'SLC22A3', 'SLC25A43', 'SLC26A4', 'SLC29A4', 'SLC2A13', 'SLC30A8', 'SLC38A1', 'SLC38A2', 'SLC38A5', 'SLC39A14', 'SLC39A5', 'SLC40A1', 'SLC43A3', 'SLC44A2', 'SLC45A3', 'SLC4A4', 'SLC4A5', 'SL
#> ℹ [2025-07-26 07:18:25] Data 3/5 of the srt_list has been log-normalized.
#> ℹ [2025-07-26 07:18:25] Perform FindVariableFeatures on the data 3/5 of the srt_list...
#> Warning: The following features were labelled as variable in 'var.features' but had no corresponding rank in `var.features.rank` and will therefore be ignored: 'ABCC3', 'ABCC8', 'ABCC9', 'ABCG1', 'ABCG2', 'ABHD2', 'ABHD4', 'ABI3', 'ACHE', 'ACP5', 'ACSL4', 'ACSS1', 'ACTN1', 'ACTN4', 'ADAM8', 'ADAM9', 'ADAMTS5', 'ADAMTS9', 'ADAMTSL2', 'ADAP2', 'ADCY1', 'ADCY3', 'ADCY5', 'ADM2', 'ADRA2A', 'ADRBK2', 'AEN', 'AGR2', 'AGRN', 'AGTRAP', 'AHNAK', 'AHR', 'AKAP7', 'ALDH1A1', 'ALDH1A2', 'ALDH1B1', 'ALOX5AP', 'ALS2CL', 'AMOTL2', 'AMT', 'ANK1', 'ANKRD1', 'ANKRD37', 'ANLN', 'ANO6', 'ANXA13', 'ANXA3', 'ANXA9', 'AP5Z1', 'AQP3', 'ARC', 'ARG2', 'ARHGAP22', 'ARHGAP23', 'ARHGAP29', 'ARHGEF10L', 'ARHGEF2', 'ARHGEF39', 'ARHGEF40', 'ARL14', 'ARL4C', 'ARL6IP1', 'ARNTL2', 'ARX', 'ASAP1', 'ASAP2', 'ASCL2', 'ASF1B', 'ASGR1', 'ASNS', 'ASRGL1', 'ASS1', 'ATF3', 'ATF5', 'ATP11A', 'ATP1A1', 'ATP1B1', 'ATP1B2', 'ATP2A3', 'ATP8A1', 'B4GALT1', 'B4GALT5', 'BACE2', 'BAIAP2L1', 'BAIAP3', 'BAMBI', 'BBC3', 'BCL2L15', 'BCL3', 'BHLHE40', 'BHLHE41', 'BIK', 'BIRC5', 'BLNK', 'BMF', 'BMP1', 'BMP2', 'BMP7', 'BST2', 'BUB1B', 'BUB1', 'C1QA', 'C1QC', 'C1QL1', 'C2CD4A', 'C2CD4B', 'C2', 'CACNA1A', 'CACNA2D1', 'CACNG4', 'CADM2', 'CADPS', 'CALY', 'CAMK2B', 'CAPG', 'CAPN5', 'CAPN6', 'CARTPT', 'CASP9', 'CAV2', 'CBLB', 'CBLN4', 'CBS', 'CCBE1', 'CCDC34', 'CCDC51', 'CCDC69', 'CCNA1', 'CD14', 'CD276', 'CD300A', 'CD68', 'CD82', 'CD9', 'CDC20', 'CDC42EP3', 'CDCA8', 'CDH17', 'CDH3', 'CDK1', 'CDK5R2', 'CDKN1A', 'CDKN2A', 'CDKN2B', 'CDKN3', 'CDR2L', 'CDX2', 'CEBPB', 'CELF3', 'CENPW', 'CERCAM', 'CFI', 'CHAC1', 'CHDH', 'CHGB', 'CHRNA3', 'CITED2', 'CITED4', 'CKAP2', 'CKAP5', 'CKMT2', 'CKS1B', 'CKS2', 'CLCF1', 'CLDN1', 'CLIC4', 'CLMN', 'CLSPN', 'CMPK2', 'CMTM7', 'CNIH2', 'CNKSR2', 'CNN2', 'CNN3', 'CNTN1', 'COL13A1', 'COL16A1', 'COL18A1', 'COL4A2', 'COL5A3', 'COL9A2', 'COPG1', 'COPZ2', 'CORO1C', 'COTL1', 'COX7A1', 'CPLX2', 'CPPED1', 'CPVL', 'CRABP2', 'CRAT', 'CREB3L1', 'CREB5', 'CRIP1', 'CRLF1', 'CRTAC1', 'CRTAP', 'CRYM', 'CSF1R', 'CSGALNACT1', 'CTSC', 'CTSH', 'CTSO', 'CTSZ', 'CWC25', 'CX3CL1', 'CXADR', 'CXCL12', 'CXCL16', 'CXCL1', 'CYB5A', 'CYP26B1', 'CYP27A1', 'CYTH3', 'DAB2IP', 'DACH1', 'DACT1', 'DAPP1', 'DBN1', 'DBNDD1', 'DCUN1D2', 'DDC', 'DDIT4', 'DDX51', 'DEDD2', 'DHODH', 'DHRS2', 'DIP2A', 'DIRAS2', 'DNAJC12', 'DNAL1', 'DOCK4', 'DOCK6', 'DOCK8', 'DOCK9', 'DOK5', 'DPP4', 'DPP6', 'DSC2', 'DSEL', 'DUSP1', 'DUSP23', 'DUSP26', 'DUSP5', 'ECEL1', 'ECM1', 'ECT2', 'EDN2', 'EEF1D', 'EEF2K', 'EFEMP2', 'EFHD2', 'EFNA1', 'EGFR', 'EGR1', 'EGR4', 'EHD2', 'EHD4', 'EHF', 'ELAVL4', 'EMILIN1', 'EMP3', 'ENAH', 'ENO2', 'ENPP2', 'EPHB2', 'EPHB4', 'EPS8', 'ERBB3', 'ERO1L', 'ERRFI1', 'ETS2', 'EVA1A', 'EVA1B', 'F2RL2', 'F2RL3', 'F2R', 'F5', 'FA2H', 'FAM105A', 'FAM107B', 'FAM129B', 'FAM134B', 'FAM171A1', 'FAM227A', 'FAM43A', 'FAM46A', 'FAM46C', 'FAM84A', 'FBLIM1', 'FBP1', 'FBXL20', 'FBXO2', 'FBXO32', 'FBXO5', 'FBXO6', 'FCER1G', 'FCGRT', 'FERMT1', 'FEV', 'FFAR4', 'FGB', 'FGF13', 'FGFR2', 'FGFR3', 'FHL2', 'FHOD1', 'FJX1', 'FKBP10', 'FKBP11', 'FKBP5', 'FLNA', 'FMNL3', 'FNDC4', 'FNIP2', 'FOSB', 'FOSL2', 'FOXA3', 'FOXC1', 'FOXM1', 'FOXQ1', 'FRZB', 'FSCN1', 'FSTL3', 'FTH1', 'FURIN', 'FUT8', 'FXYD3', 'FXYD5', 'FXYD6', 'G0S2', 'G6PC', 'GAD2', 'GADD45A', 'GADD45B', 'GALE', 'GALM', 'GALNT2', 'GAL', 'GAS6', 'GAS7', 'GATA2', 'GATA4', 'GATSL3', 'GCGR', 'GCH1', 'GCK', 'GCLC', 'GCNT1', 'GCNT3', 'GC', 'GDA', 'GEM', 'GFOD2', 'GFPT2', 'GFRA1', 'GJA1', 'GJB1', 'GK', 'GLIPR1', 'GLRX', 'GLS', 'GMNN', 'GNPAT', 'GOLM1', 'GPC1', 'GPC6', 'GPD1', 'GPR160', 'GPR161', 'GPSM1', 'GPX3', 'GPX8', 'GRASP', 'GREB1', 'GSN', 'GSTM5', 'GSTP1', 'GTF3C3', 'GTPBP10', 'GTSE1', 'GULP1', 'H19', 'HABP2', 'HAND2', 'HDAC9', 'HEPACAM2', 'HEPH', 'HERPUD1', 'HES1', 'HEYL', 'HHEX', 'HIF1A', 'HIF3A', 'HILPDA', 'HIST1H1C', 'HIST1H2BK', 'HIST2H2BE', 'HK1', 'HK2', 'HKDC1', 'HMGB2', 'HMGB3', 'HMGCR', 'HMGCS1', 'HMMR', 'HN1', 'HNF1B', 'HOGA1', 'HOMER3', 'HOPX', 'HOXB2', 'HPCAL1', 'HPGD', 'HPN', 'HS3ST1', 'HSD11B2', 'HSD17B11', 'HSP90B1', 'HSPB1', 'HSPB8', 'HSPH1', 'ID1', 'ID4', 'IDH1', 'IDH2', 'IER2', 'IER3', 'IFIT1', 'IFITM2', 'IFNLR1', 'IGSF10', 'IGSF1', 'IGSF3', 'IL11', 'IL18', 'IL1R2', 'IMPA2', 'INPP5F', 'INSIG1', 'INSM1', 'IP6K1', 'IPCEF1', 'IRAK4', 'IRF1', 'IRF8', 'IRS2', 'IRX1', 'IRX2', 'IRX4', 'ISL1', 'ITGA11', 'ITGA3', 'ITGA9', 'ITGAV', 'ITGB4', 'ITGB6', 'ITGB8', 'ITIH5', 'ITPRIPL2', 'JAG1', 'JARID2', 'JRK', 'JSRP1', 'JUN', 'KANK1', 'KANK2', 'KCNA5', 'KCNAB1', 'KCNE3', 'KCNG1', 'KCNH6', 'KCNJ11', 'KCNJ16', 'KCNJ5', 'KCNJ6', 'KCNK16', 'KCNK3', 'KCNK5', 'KCNMA1', 'KCNN3', 'KCNQ1OT1', 'KCNQ1', 'KCTD12', 'KCTD8', 'KDELC2', 'KDELR3', 'KDM5D', 'KIF11', 'KIF12', 'KIF14', 'KIF21B', 'KIF2C', 'KIF5C', 'KISS1R', 'KLF2', 'KLF4', 'KLF5', 'KLF6', 'KLHDC8A', 'KLHL21', 'KLK11', 'KNSTRN', 'KPNA2', 'KRT17', 'KRTCAP3', 'L2HGDH', 'LAD1', 'LAMB1', 'LAMB2', 'LAPTM5', 'LBH', 'LDLR', 'LECT1', 'LEFTY1', 'LGALS3', 'LGALS9', 'LGI2', 'LHFP', 'LIMA1', 'LIMCH1', 'LIMK2', 'LIMS1', 'LIPH', 'LMO7', 'LONRF2', 'LOXL2', 'LPAR6', 'LRP12', 'LRP1', 'LRRC10B', 'LRRC59', 'LSAMP', 'LTBP4', 'LURAP1L', 'LUZP1', 'LXN', 'LY6E', 'LY6H', 'LYN', 'LYPD1', 'LYPD6B', 'MACC1', 'MAFA', 'MAFB', 'MAFF', 'MAL2', 'MALL', 'MAN1C1', 'MANF', 'MAOB', 'MAP1B', 'MAP2', 'MAP7D2', 'MAPT', 'MARCKSL1', 'MATN2', 'MDFI', 'MDK', 'MECOM', 'MEIS2', 'MERTK', 'METRN', 'METTL20', 'METTL22', 'MFI2', 'MGAT4A', 'MGAT4B', 'MGLL', 'MICAL2', 'MID1IP1', 'MIR143HG', 'MLLT11', 'MLXIPL', 'MMP14', 'MNX1', 'MPZL2', 'MRC1', 'MRC2', 'MSMO1', 'MSN', 'MSRB1', 'MSRB3', 'MTUS1', 'MUM1L1', 'MYC', 'MYH10', 'MYH9', 'MYL12A', 'MYO10', 'MYO1E', 'MYO5B', 'MYRF', 'MYZAP', 'NAA16', 'NCAM1', 'NCAPG', 'NCAPH', 'NCMAP', 'NDRG1', 'NDRG2', 'NDRG4', 'NDUFA4L2', 'NDUFA4', 'NEK8', 'NEURL1B', 'NEURL3', 'NEUROD1', 'NFATC4', 'NFKBIA', 'NFKBIE', 'NINJ1', 'NKD1', 'NKX2-2', 'NLN', 'NMB', 'NMNAT1', 'NOB1', 'NOTCH1', 'NOTCH2', 'NOTCH3', 'NOTCH4', 'NOX4', 'NPHS1', 'NPNT', 'NPTX2', 'NQO1', 'NR0B1', 'NR0B2', 'NR1D1', 'NR2F2', 'NR4A2', 'NR5A2', 'NRP2', 'NTN4', 'NUAK2', 'NUF2', 'NUSAP1', 'OCLN', 'OGDHL', 'OLFML2B', 'OPA3', 'ORM1', 'P4HA3', 'PACSIN2', 'PAH', 'PAK3', 'PALLD', 'PAMR1', 'PAPSS2', 'PARM1', 'PAX6', 'PBK', 'PCDH17', 'PCDH18', 'PCDH9', 'PCK1', 'PCNA', 'PCP4', 'PCSK2', 'PCSK5', 'PDE3A', 'PDE3B', 'PDE8B', 'PDGFA', 'PDGFD', 'PDIA4', 'PDLIM1', 'PDLIM4', 'PDLIM7', 'PDP2', 'PDX1', 'PEMT', 'PER3', 'PEX13', 'PFKFB3', 'PGBD5', 'PGM1', 'PGM2L1', 'PHF19', 'PHGDH', 'PHGR1', 'PHLDA2', 'PHLDB1', 'PID1', 'PIEZO1', 'PIK3AP1', 'PIK3R1', 'PIM1', 'PKIB', 'PKNOX2', 'PKP2', 'PLA2G4C', 'PLA2G7', 'PLAA', 'PLAGL1', 'PLAT', 'PLCXD3', 'PLEKHA2', 'PLEKHB1', 'PLEKHG2', 'PLEKHO1', 'PLIN5', 'PLK2', 'PLLP', 'PLOD2', 'PLSCR1', 'PLXDC1', 'PLXNA1', 'PLXNA2', 'PLXND1', 'PMAIP1', 'PNMA2', 'PNRC1', 'POC1A', 'POLD4', 'PON2', 'POU2F2', 'PPARGC1A', 'PPIC', 'PPIF', 'PPL', 'PPP1R15A', 'PPP1R18', 'PPP1R1A', 'PPP1R1B', 'PPP2R2B', 'PRC1', 'PRELP', 'PRKCDBP', 'PRKG1', 'PRKX', 'PRLHR', 'PRMT6', 'PRR11', 'PRR15L', 'PRSS22', 'PRSS23', 'PRSS8', 'PTAFR', 'PTGDS', 'PTGES', 'PTGFR', 'PTK7', 'PTPN14', 'PTPN3', 'PTPRN', 'PTPRT', 'PTRF', 'PTTG1', 'PVRL3', 'PVRL4', 'PYCARD', 'PYCR1', 'PYROXD2', 'QPRT', 'QSOX1', 'RAB11FIP1', 'RAB33A', 'RAB3B', 'RAB42', 'RACGAP1', 'RAMP1', 'RAP1GAP', 'RAPGEF5', 'RARG', 'RARRES1', 'RASA3', 'RASEF', 'RASGRP3', 'RASL12', 'RASSF4', 'RASSF9', 'RBM38', 'RBP1', 'RBPMS', 'RCL1', 'RCN3', 'RDH10', 'RDX', 'RELB', 'RETSAT', 'RFTN1', 'RFX6', 'RGS5', 'RHBDF1', 'RHOB', 'RHOC', 'RHOD', 'RHOV', 'RHPN2', 'RINL', 'RNASE6', 'RNF114', 'RNF157', 'RNF24', 'RNF32', 'ROBO1', 'ROR2', 'RPL22L1', 'RPS6KA2', 'RRAGD', 'RSU1', 'S100A10', 'S100A11', 'S100A16', 'S1PR3', 'SAMHD1', 'SCG2', 'SCG3', 'SCN1B', 'SCNN1A', 'SCTR', 'SDC2', 'SDS', 'SEL1L3', 'SEMA3B', 'SEMA3C', 'SEMA4B', 'SEMA6A', 'SEMA7A', 'SEPT3', 'SEPT6', 'SEPT9', 'SERPINA6', 'SERPINE2', 'SERPINF1', 'SERPINI1', 'SERTM1', 'SETX', 'SEZ6L', 'SFMBT2', 'SFN', 'SFRP1', 'SFRP5', 'SGOL1', 'SGSM1', 'SGSM3', 'SH3BGRL3', 'SH3BP4', 'SH3KBP1', 'SH3PXD2A', 'SHANK2', 'SHC1', 'SHROOM1', 'SHROOM3', 'SIRPA', 'SIX4', 'SKIL', 'SLC12A2', 'SLC12A7', 'SLC15A4', 'SLC16A3', 'SLC16A5', 'SLC16A7', 'SLC16A9', 'SLC1A5', 'SLC22A17', 'SLC22A3', 'SLC25A43', 'SLC26A4', 'SLC29A4', 'SLC2A13', 'SLC30A8', 'SLC38A1', 'SLC38A2', 'SLC38A5', 'SLC39A14', 'SLC39A5', 'SLC40A1', 'SLC43A3', 'SLC44A2', 'SLC45A3', 'SLC4A4', 'SLC4A5', 'SL
#> ℹ [2025-07-26 07:18:26] Data 4/5 of the srt_list has been log-normalized.
#> ℹ [2025-07-26 07:18:26] Perform FindVariableFeatures on the data 4/5 of the srt_list...
#> Warning: The following features were labelled as variable in 'var.features' but had no corresponding rank in `var.features.rank` and will therefore be ignored: 'ABCC3', 'ABCC8', 'ABCC9', 'ABCG1', 'ABCG2', 'ABHD2', 'ABHD4', 'ABI3', 'ACHE', 'ACP5', 'ACSL4', 'ACSS1', 'ACTN1', 'ACTN4', 'ADAM8', 'ADAM9', 'ADAMTS5', 'ADAMTS9', 'ADAMTSL2', 'ADAP2', 'ADCY1', 'ADCY3', 'ADCY5', 'ADM2', 'ADRA2A', 'ADRBK2', 'AEN', 'AGR2', 'AGRN', 'AGTRAP', 'AHNAK', 'AHR', 'AKAP7', 'ALDH1A1', 'ALDH1A2', 'ALDH1B1', 'ALOX5AP', 'ALS2CL', 'AMOTL2', 'AMT', 'ANK1', 'ANKRD1', 'ANKRD37', 'ANLN', 'ANO6', 'ANXA13', 'ANXA3', 'ANXA9', 'AP5Z1', 'AQP3', 'ARC', 'ARG2', 'ARHGAP22', 'ARHGAP23', 'ARHGAP29', 'ARHGEF10L', 'ARHGEF2', 'ARHGEF39', 'ARHGEF40', 'ARL14', 'ARL4C', 'ARL6IP1', 'ARNTL2', 'ARX', 'ASAP1', 'ASAP2', 'ASCL2', 'ASF1B', 'ASGR1', 'ASNS', 'ASRGL1', 'ASS1', 'ATF3', 'ATF5', 'ATP11A', 'ATP1A1', 'ATP1B1', 'ATP1B2', 'ATP2A3', 'ATP8A1', 'B4GALT1', 'B4GALT5', 'BACE2', 'BAIAP2L1', 'BAIAP3', 'BAMBI', 'BBC3', 'BCL2L15', 'BCL3', 'BHLHE40', 'BHLHE41', 'BIK', 'BIRC5', 'BLNK', 'BMF', 'BMP1', 'BMP2', 'BMP7', 'BST2', 'BUB1B', 'BUB1', 'C1QA', 'C1QC', 'C1QL1', 'C2CD4A', 'C2CD4B', 'C2', 'CACNA1A', 'CACNA2D1', 'CACNG4', 'CADM2', 'CADPS', 'CALY', 'CAMK2B', 'CAPG', 'CAPN5', 'CAPN6', 'CARTPT', 'CASP9', 'CAV2', 'CBLB', 'CBLN4', 'CBS', 'CCBE1', 'CCDC34', 'CCDC51', 'CCDC69', 'CCNA1', 'CD14', 'CD276', 'CD300A', 'CD68', 'CD82', 'CD9', 'CDC20', 'CDC42EP3', 'CDCA8', 'CDH17', 'CDH3', 'CDK1', 'CDK5R2', 'CDKN1A', 'CDKN2A', 'CDKN2B', 'CDKN3', 'CDR2L', 'CDX2', 'CEBPB', 'CELF3', 'CENPW', 'CERCAM', 'CFI', 'CHAC1', 'CHDH', 'CHGB', 'CHRNA3', 'CITED2', 'CITED4', 'CKAP2', 'CKAP5', 'CKMT2', 'CKS1B', 'CKS2', 'CLCF1', 'CLDN1', 'CLIC4', 'CLMN', 'CLSPN', 'CMPK2', 'CMTM7', 'CNIH2', 'CNKSR2', 'CNN2', 'CNN3', 'CNTN1', 'COL13A1', 'COL16A1', 'COL18A1', 'COL4A2', 'COL5A3', 'COL9A2', 'COPG1', 'COPZ2', 'CORO1C', 'COTL1', 'COX7A1', 'CPLX2', 'CPPED1', 'CPVL', 'CRABP2', 'CRAT', 'CREB3L1', 'CREB5', 'CRIP1', 'CRLF1', 'CRTAC1', 'CRTAP', 'CRYM', 'CSF1R', 'CSGALNACT1', 'CTSC', 'CTSH', 'CTSO', 'CTSZ', 'CWC25', 'CX3CL1', 'CXADR', 'CXCL12', 'CXCL16', 'CXCL1', 'CYB5A', 'CYP26B1', 'CYP27A1', 'CYTH3', 'DAB2IP', 'DACH1', 'DACT1', 'DAPP1', 'DBN1', 'DBNDD1', 'DCUN1D2', 'DDC', 'DDIT4', 'DDX51', 'DEDD2', 'DHODH', 'DHRS2', 'DIP2A', 'DIRAS2', 'DNAJC12', 'DNAL1', 'DOCK4', 'DOCK6', 'DOCK8', 'DOCK9', 'DOK5', 'DPP4', 'DPP6', 'DSC2', 'DSEL', 'DUSP1', 'DUSP23', 'DUSP26', 'DUSP5', 'ECEL1', 'ECM1', 'ECT2', 'EDN2', 'EEF1D', 'EEF2K', 'EFEMP2', 'EFHD2', 'EFNA1', 'EGFR', 'EGR1', 'EGR4', 'EHD2', 'EHD4', 'EHF', 'ELAVL4', 'EMILIN1', 'EMP3', 'ENAH', 'ENO2', 'ENPP2', 'EPHB2', 'EPHB4', 'EPS8', 'ERBB3', 'ERO1L', 'ERRFI1', 'ETS2', 'EVA1A', 'EVA1B', 'F2RL2', 'F2RL3', 'F2R', 'F5', 'FA2H', 'FAM105A', 'FAM107B', 'FAM129B', 'FAM134B', 'FAM171A1', 'FAM227A', 'FAM43A', 'FAM46A', 'FAM46C', 'FAM84A', 'FBLIM1', 'FBP1', 'FBXL20', 'FBXO2', 'FBXO32', 'FBXO5', 'FBXO6', 'FCER1G', 'FCGRT', 'FERMT1', 'FEV', 'FFAR4', 'FGB', 'FGF13', 'FGFR2', 'FGFR3', 'FHL2', 'FHOD1', 'FJX1', 'FKBP10', 'FKBP11', 'FKBP5', 'FLNA', 'FMNL3', 'FNDC4', 'FNIP2', 'FOSB', 'FOSL2', 'FOXA3', 'FOXC1', 'FOXM1', 'FOXQ1', 'FRZB', 'FSCN1', 'FSTL3', 'FTH1', 'FURIN', 'FUT8', 'FXYD3', 'FXYD5', 'FXYD6', 'G0S2', 'G6PC', 'GAD2', 'GADD45A', 'GADD45B', 'GALE', 'GALM', 'GALNT2', 'GAL', 'GAS6', 'GAS7', 'GATA2', 'GATA4', 'GATSL3', 'GCGR', 'GCH1', 'GCK', 'GCLC', 'GCNT1', 'GCNT3', 'GC', 'GDA', 'GEM', 'GFOD2', 'GFPT2', 'GFRA1', 'GJA1', 'GJB1', 'GK', 'GLIPR1', 'GLRX', 'GLS', 'GMNN', 'GNPAT', 'GOLM1', 'GPC1', 'GPC6', 'GPD1', 'GPR160', 'GPR161', 'GPSM1', 'GPX3', 'GPX8', 'GRASP', 'GREB1', 'GSN', 'GSTM5', 'GSTP1', 'GTF3C3', 'GTPBP10', 'GTSE1', 'GULP1', 'H19', 'HABP2', 'HAND2', 'HDAC9', 'HEPACAM2', 'HEPH', 'HERPUD1', 'HES1', 'HEYL', 'HHEX', 'HIF1A', 'HIF3A', 'HILPDA', 'HIST1H1C', 'HIST1H2BK', 'HIST2H2BE', 'HK1', 'HK2', 'HKDC1', 'HMGB2', 'HMGB3', 'HMGCR', 'HMGCS1', 'HMMR', 'HN1', 'HNF1B', 'HOGA1', 'HOMER3', 'HOPX', 'HOXB2', 'HPCAL1', 'HPGD', 'HPN', 'HS3ST1', 'HSD11B2', 'HSD17B11', 'HSP90B1', 'HSPB1', 'HSPB8', 'HSPH1', 'ID1', 'ID4', 'IDH1', 'IDH2', 'IER2', 'IER3', 'IFIT1', 'IFITM2', 'IFNLR1', 'IGSF10', 'IGSF1', 'IGSF3', 'IL11', 'IL18', 'IL1R2', 'IMPA2', 'INPP5F', 'INSIG1', 'INSM1', 'IP6K1', 'IPCEF1', 'IRAK4', 'IRF1', 'IRF8', 'IRS2', 'IRX1', 'IRX2', 'IRX4', 'ISL1', 'ITGA11', 'ITGA3', 'ITGA9', 'ITGAV', 'ITGB4', 'ITGB6', 'ITGB8', 'ITIH5', 'ITPRIPL2', 'JAG1', 'JARID2', 'JRK', 'JSRP1', 'JUN', 'KANK1', 'KANK2', 'KCNA5', 'KCNAB1', 'KCNE3', 'KCNG1', 'KCNH6', 'KCNJ11', 'KCNJ16', 'KCNJ5', 'KCNJ6', 'KCNK16', 'KCNK3', 'KCNK5', 'KCNMA1', 'KCNN3', 'KCNQ1OT1', 'KCNQ1', 'KCTD12', 'KCTD8', 'KDELC2', 'KDELR3', 'KDM5D', 'KIF11', 'KIF12', 'KIF14', 'KIF21B', 'KIF2C', 'KIF5C', 'KISS1R', 'KLF2', 'KLF4', 'KLF5', 'KLF6', 'KLHDC8A', 'KLHL21', 'KLK11', 'KNSTRN', 'KPNA2', 'KRT17', 'KRTCAP3', 'L2HGDH', 'LAD1', 'LAMB1', 'LAMB2', 'LAPTM5', 'LBH', 'LDLR', 'LECT1', 'LEFTY1', 'LGALS3', 'LGALS9', 'LGI2', 'LHFP', 'LIMA1', 'LIMCH1', 'LIMK2', 'LIMS1', 'LIPH', 'LMO7', 'LONRF2', 'LOXL2', 'LPAR6', 'LRP12', 'LRP1', 'LRRC10B', 'LRRC59', 'LSAMP', 'LTBP4', 'LURAP1L', 'LUZP1', 'LXN', 'LY6E', 'LY6H', 'LYN', 'LYPD1', 'LYPD6B', 'MACC1', 'MAFA', 'MAFB', 'MAFF', 'MAL2', 'MALL', 'MAN1C1', 'MANF', 'MAOB', 'MAP1B', 'MAP2', 'MAP7D2', 'MAPT', 'MARCKSL1', 'MATN2', 'MDFI', 'MDK', 'MECOM', 'MEIS2', 'MERTK', 'METRN', 'METTL20', 'METTL22', 'MFI2', 'MGAT4A', 'MGAT4B', 'MGLL', 'MICAL2', 'MID1IP1', 'MIR143HG', 'MLLT11', 'MLXIPL', 'MMP14', 'MNX1', 'MPZL2', 'MRC1', 'MRC2', 'MSMO1', 'MSN', 'MSRB1', 'MSRB3', 'MTUS1', 'MUM1L1', 'MYC', 'MYH10', 'MYH9', 'MYL12A', 'MYO10', 'MYO1E', 'MYO5B', 'MYRF', 'MYZAP', 'NAA16', 'NCAM1', 'NCAPG', 'NCAPH', 'NCMAP', 'NDRG1', 'NDRG2', 'NDRG4', 'NDUFA4L2', 'NDUFA4', 'NEK8', 'NEURL1B', 'NEURL3', 'NEUROD1', 'NFATC4', 'NFKBIA', 'NFKBIE', 'NINJ1', 'NKD1', 'NKX2-2', 'NLN', 'NMB', 'NMNAT1', 'NOB1', 'NOTCH1', 'NOTCH2', 'NOTCH3', 'NOTCH4', 'NOX4', 'NPHS1', 'NPNT', 'NPTX2', 'NQO1', 'NR0B1', 'NR0B2', 'NR1D1', 'NR2F2', 'NR4A2', 'NR5A2', 'NRP2', 'NTN4', 'NUAK2', 'NUF2', 'NUSAP1', 'OCLN', 'OGDHL', 'OLFML2B', 'OPA3', 'ORM1', 'P4HA3', 'PACSIN2', 'PAH', 'PAK3', 'PALLD', 'PAMR1', 'PAPSS2', 'PARM1', 'PAX6', 'PBK', 'PCDH17', 'PCDH18', 'PCDH9', 'PCK1', 'PCNA', 'PCP4', 'PCSK2', 'PCSK5', 'PDE3A', 'PDE3B', 'PDE8B', 'PDGFA', 'PDGFD', 'PDIA4', 'PDLIM1', 'PDLIM4', 'PDLIM7', 'PDP2', 'PDX1', 'PEMT', 'PER3', 'PEX13', 'PFKFB3', 'PGBD5', 'PGM1', 'PGM2L1', 'PHF19', 'PHGDH', 'PHGR1', 'PHLDA2', 'PHLDB1', 'PID1', 'PIEZO1', 'PIK3AP1', 'PIK3R1', 'PIM1', 'PKIB', 'PKNOX2', 'PKP2', 'PLA2G4C', 'PLA2G7', 'PLAA', 'PLAGL1', 'PLAT', 'PLCXD3', 'PLEKHA2', 'PLEKHB1', 'PLEKHG2', 'PLEKHO1', 'PLIN5', 'PLK2', 'PLLP', 'PLOD2', 'PLSCR1', 'PLXDC1', 'PLXNA1', 'PLXNA2', 'PLXND1', 'PMAIP1', 'PNMA2', 'PNRC1', 'POC1A', 'POLD4', 'PON2', 'POU2F2', 'PPARGC1A', 'PPIC', 'PPIF', 'PPL', 'PPP1R15A', 'PPP1R18', 'PPP1R1A', 'PPP1R1B', 'PPP2R2B', 'PRC1', 'PRELP', 'PRKCDBP', 'PRKG1', 'PRKX', 'PRLHR', 'PRMT6', 'PRR11', 'PRR15L', 'PRSS22', 'PRSS23', 'PRSS8', 'PTAFR', 'PTGDS', 'PTGES', 'PTGFR', 'PTK7', 'PTPN14', 'PTPN3', 'PTPRN', 'PTPRT', 'PTRF', 'PTTG1', 'PVRL3', 'PVRL4', 'PYCARD', 'PYCR1', 'PYROXD2', 'QPRT', 'QSOX1', 'RAB11FIP1', 'RAB33A', 'RAB3B', 'RAB42', 'RACGAP1', 'RAMP1', 'RAP1GAP', 'RAPGEF5', 'RARG', 'RARRES1', 'RASA3', 'RASEF', 'RASGRP3', 'RASL12', 'RASSF4', 'RASSF9', 'RBM38', 'RBP1', 'RBPMS', 'RCL1', 'RCN3', 'RDH10', 'RDX', 'RELB', 'RETSAT', 'RFTN1', 'RFX6', 'RGS5', 'RHBDF1', 'RHOB', 'RHOC', 'RHOD', 'RHOV', 'RHPN2', 'RINL', 'RNASE6', 'RNF114', 'RNF157', 'RNF24', 'RNF32', 'ROBO1', 'ROR2', 'RPL22L1', 'RPS6KA2', 'RRAGD', 'RSU1', 'S100A10', 'S100A11', 'S100A16', 'S1PR3', 'SAMHD1', 'SCG2', 'SCG3', 'SCN1B', 'SCNN1A', 'SCTR', 'SDC2', 'SDS', 'SEL1L3', 'SEMA3B', 'SEMA3C', 'SEMA4B', 'SEMA6A', 'SEMA7A', 'SEPT3', 'SEPT6', 'SEPT9', 'SERPINA6', 'SERPINE2', 'SERPINF1', 'SERPINI1', 'SERTM1', 'SETX', 'SEZ6L', 'SFMBT2', 'SFN', 'SFRP1', 'SFRP5', 'SGOL1', 'SGSM1', 'SGSM3', 'SH3BGRL3', 'SH3BP4', 'SH3KBP1', 'SH3PXD2A', 'SHANK2', 'SHC1', 'SHROOM1', 'SHROOM3', 'SIRPA', 'SIX4', 'SKIL', 'SLC12A2', 'SLC12A7', 'SLC15A4', 'SLC16A3', 'SLC16A5', 'SLC16A7', 'SLC16A9', 'SLC1A5', 'SLC22A17', 'SLC22A3', 'SLC25A43', 'SLC26A4', 'SLC29A4', 'SLC2A13', 'SLC30A8', 'SLC38A1', 'SLC38A2', 'SLC38A5', 'SLC39A14', 'SLC39A5', 'SLC40A1', 'SLC43A3', 'SLC44A2', 'SLC45A3', 'SLC4A4', 'SLC4A5', 'SL
#> ℹ [2025-07-26 07:18:26] Data 5/5 of the srt_list has been log-normalized.
#> ℹ [2025-07-26 07:18:26] Perform FindVariableFeatures on the data 5/5 of the srt_list...
#> ℹ [2025-07-26 07:18:27] Use the separate HVF from srt_list...
#> ℹ [2025-07-26 07:18:27] Number of available HVF: 259
#> ℹ [2025-07-26 07:18:27] Finished checking.
#> ℹ [2025-07-26 07:18:29] Perform integration(Uncorrected) on the data...
#> ℹ [2025-07-26 07:18:29] Perform ScaleData on the data...
#> ℹ [2025-07-26 07:18:29] Perform linear dimension reduction (pca) on the data...
#> ℹ [2025-07-26 07:18:29] Linear_reduction(pca) is already existed. Skip calculation.
#> Warning: The following arguments are not used: features
#> Warning: multiple layers are identified by counts.1.1.1.1 counts.2.1 counts.2.1.1 counts.2.1.1.1 data.1.1.1.1 scale.data.1.1.1.1 data.2.1.1.1 scale.data.2.1.1.1 data.2.1.1 scale.data.2.1.1 data.2.1 scale.data.2.1
#> only the first layer is used
#> ! [2025-07-26 07:18:29] Error: Not compatible with requested type: [type=S4; target=double].
#> ! [2025-07-26 07:18:29] Error when performing FindClusters. Skip this step...
#> ℹ [2025-07-26 07:18:29] Perform nonlinear dimension reduction (umap) on the data...
#> Warning: no non-missing arguments to min; returning Inf
#> Warning: no non-missing arguments to max; returning -Inf
#> ℹ [2025-07-26 07:18:29] Non-linear dimensionality reduction(umap) using Reduction(Uncorrectedpca, dims:Inf--Inf) as input
#> ! [2025-07-26 07:18:29] Error in `object[[graph]]`:
#> ! [2025-07-26 07:18:29] ! ‘Uncorrected_SNN’ not found in this Seurat object
#> ! [2025-07-26 07:18:29]
#> ! [2025-07-26 07:18:29] Error when performing nonlinear dimension reduction. Skip this step...
#> ℹ [2025-07-26 07:18:31] Uncorrected_integrate done
#> ℹ [2025-07-26 07:18:31] Elapsed time: 8.71 secs
CellDimPlot(
panc8_sub,
group.by = c("tech", "celltype")
)
#> Warning: No shared levels found between `names(values)` of the manual scale and the
#> data's fill values.
#> Warning: No shared levels found between `names(values)` of the manual scale and the
#> data's fill values.
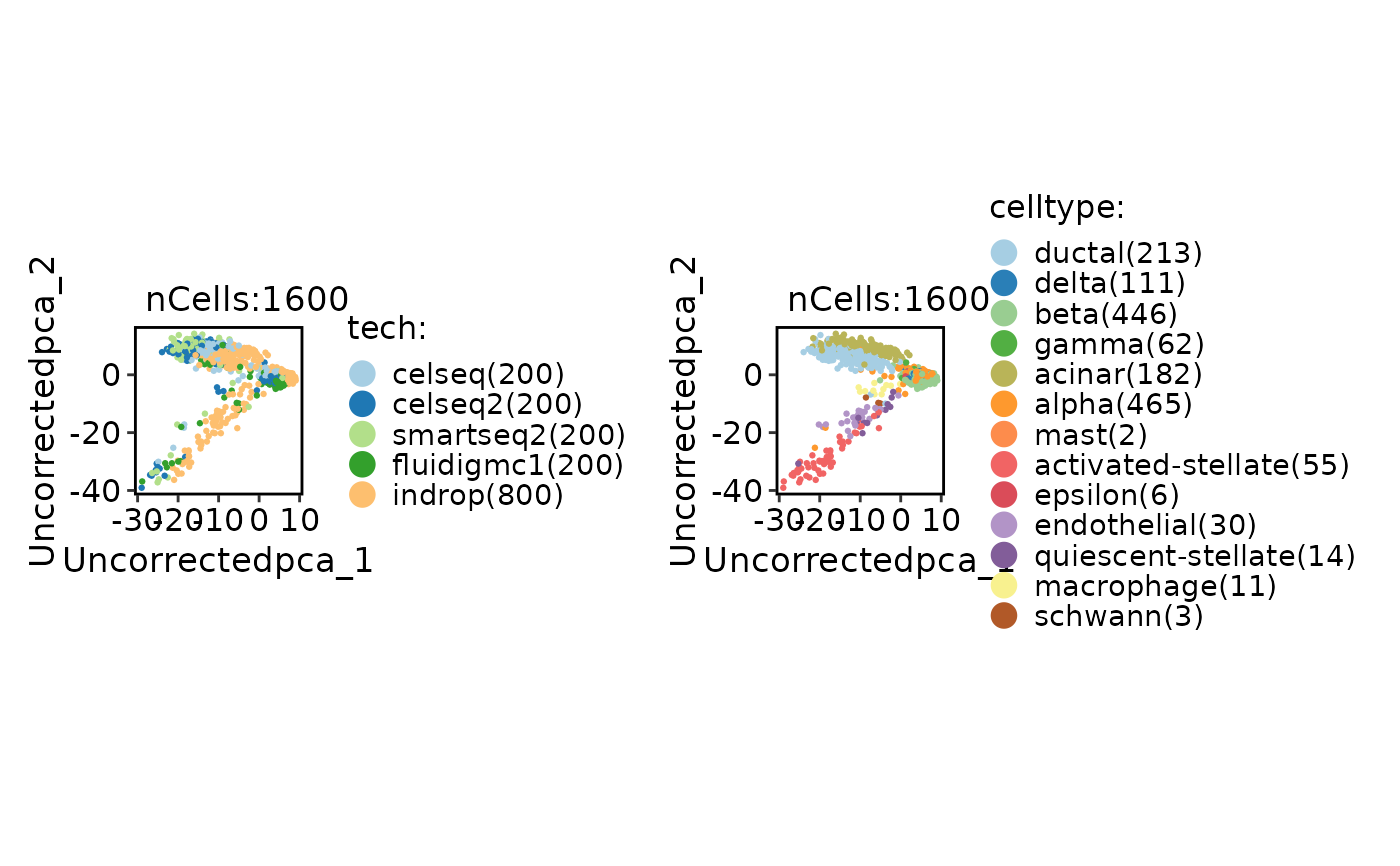 panc8_sub <- integration_scop(
panc8_sub,
batch = "tech",
integration_method = "Uncorrected",
HVF_min_intersection = 5,
scale_within_batch = TRUE
)
#> ℹ [2025-07-26 07:18:31] Start Uncorrected_integrate
#> ℹ [2025-07-26 07:18:31] Spliting srt_merge into srt_list by column tech...
#> ℹ [2025-07-26 07:18:33] Checking srt_list...
#> Warning: The following features were labelled as variable in 'var.features' but had no corresponding rank in `var.features.rank` and will therefore be ignored: 'AHNAK', 'ANXA3', 'CDKN1A', 'CFI', 'CLDN1', 'CNN3', 'CTSH', 'DUSP1', 'FAM84A', 'GCNT3', 'GC', 'ID1', 'KLF6', 'LAD1', 'MECOM', 'MSN', 'MTUS1', 'PALLD', 'PHGDH', 'PLK2', 'PPP2R2B', 'RHOC', 'S100A11', 'S100A16', 'SCTR', 'SLC43A3', 'SLC4A4', 'SOX9', 'SPOCK1', 'SYNPO', 'TC2N', 'TFPI2', 'TRIB3', 'VTCN1', 'XBP1', 'ZFP36L1'
#> ℹ [2025-07-26 07:18:33] Data 1/5 of the srt_list has been log-normalized.
#> ℹ [2025-07-26 07:18:33] Perform FindVariableFeatures on the data 1/5 of the srt_list...
#> Warning: The following features were labelled as variable in 'var.features' but had no corresponding rank in `var.features.rank` and will therefore be ignored: 'AHNAK', 'ANXA3', 'CDKN1A', 'CFI', 'CLDN1', 'CNN3', 'CTSH', 'DUSP1', 'FAM84A', 'GCNT3', 'GC', 'ID1', 'KLF6', 'LAD1', 'MECOM', 'MSN', 'MTUS1', 'PALLD', 'PHGDH', 'PLK2', 'PPP2R2B', 'RHOC', 'S100A11', 'S100A16', 'SCTR', 'SLC43A3', 'SLC4A4', 'SOX9', 'SPOCK1', 'SYNPO', 'TC2N', 'TFPI2', 'TRIB3', 'VTCN1', 'XBP1', 'ZFP36L1'
#> ℹ [2025-07-26 07:18:34] Data 2/5 of the srt_list has been log-normalized.
#> ℹ [2025-07-26 07:18:34] Perform FindVariableFeatures on the data 2/5 of the srt_list...
#> Warning: The following features were labelled as variable in 'var.features' but had no corresponding rank in `var.features.rank` and will therefore be ignored: 'AHNAK', 'ANXA3', 'CDKN1A', 'CFI', 'CLDN1', 'CNN3', 'CTSH', 'DUSP1', 'FAM84A', 'GCNT3', 'GC', 'ID1', 'KLF6', 'LAD1', 'MECOM', 'MSN', 'MTUS1', 'PALLD', 'PHGDH', 'PLK2', 'PPP2R2B', 'RHOC', 'S100A11', 'S100A16', 'SCTR', 'SLC43A3', 'SLC4A4', 'SOX9', 'SPOCK1', 'SYNPO', 'TC2N', 'TFPI2', 'TRIB3', 'VTCN1', 'XBP1', 'ZFP36L1'
#> ℹ [2025-07-26 07:18:34] Data 3/5 of the srt_list has been log-normalized.
#> ℹ [2025-07-26 07:18:34] Perform FindVariableFeatures on the data 3/5 of the srt_list...
#> Warning: The following features were labelled as variable in 'var.features' but had no corresponding rank in `var.features.rank` and will therefore be ignored: 'AHNAK', 'ANXA3', 'CDKN1A', 'CFI', 'CLDN1', 'CNN3', 'CTSH', 'DUSP1', 'FAM84A', 'GCNT3', 'GC', 'ID1', 'KLF6', 'LAD1', 'MECOM', 'MSN', 'MTUS1', 'PALLD', 'PHGDH', 'PLK2', 'PPP2R2B', 'RHOC', 'S100A11', 'S100A16', 'SCTR', 'SLC43A3', 'SLC4A4', 'SOX9', 'SPOCK1', 'SYNPO', 'TC2N', 'TFPI2', 'TRIB3', 'VTCN1', 'XBP1', 'ZFP36L1'
#> ℹ [2025-07-26 07:18:35] Data 4/5 of the srt_list has been log-normalized.
#> ℹ [2025-07-26 07:18:35] Perform FindVariableFeatures on the data 4/5 of the srt_list...
#> Warning: The following features were labelled as variable in 'var.features' but had no corresponding rank in `var.features.rank` and will therefore be ignored: 'AHNAK', 'ANXA3', 'CDKN1A', 'CFI', 'CLDN1', 'CNN3', 'CTSH', 'DUSP1', 'FAM84A', 'GCNT3', 'GC', 'ID1', 'KLF6', 'LAD1', 'MECOM', 'MSN', 'MTUS1', 'PALLD', 'PHGDH', 'PLK2', 'PPP2R2B', 'RHOC', 'S100A11', 'S100A16', 'SCTR', 'SLC43A3', 'SLC4A4', 'SOX9', 'SPOCK1', 'SYNPO', 'TC2N', 'TFPI2', 'TRIB3', 'VTCN1', 'XBP1', 'ZFP36L1'
#> ℹ [2025-07-26 07:18:35] Data 5/5 of the srt_list has been log-normalized.
#> ℹ [2025-07-26 07:18:35] Perform FindVariableFeatures on the data 5/5 of the srt_list...
#> ℹ [2025-07-26 07:18:36] Use the separate HVF from srt_list...
#> ℹ [2025-07-26 07:18:36] Number of available HVF: 259
#> ℹ [2025-07-26 07:18:36] Finished checking.
#> ℹ [2025-07-26 07:18:38] Perform integration(Uncorrected) on the data...
#> ℹ [2025-07-26 07:18:38] Perform ScaleData on the data...
#> ℹ [2025-07-26 07:18:38] Perform linear dimension reduction (pca) on the data...
#> ℹ [2025-07-26 07:18:38] Linear_reduction(pca) is already existed. Skip calculation.
#> Warning: The following arguments are not used: features
#> Warning: multiple layers are identified by counts.1.1.1.1 counts.2.1 counts.2.1.1 counts.2.1.1.1 data.1.1.1.1 scale.data.1.1.1.1 data.2.1.1.1 scale.data.2.1.1.1 data.2.1.1 scale.data.2.1.1 data.2.1 scale.data.2.1
#> only the first layer is used
#> ! [2025-07-26 07:18:38] Error: Not compatible with requested type: [type=S4; target=double].
#> ! [2025-07-26 07:18:38] Error when performing FindClusters. Skip this step...
#> ℹ [2025-07-26 07:18:38] Perform nonlinear dimension reduction (umap) on the data...
#> Warning: no non-missing arguments to min; returning Inf
#> Warning: no non-missing arguments to max; returning -Inf
#> ℹ [2025-07-26 07:18:39] Non-linear dimensionality reduction(umap) using Reduction(Uncorrectedpca, dims:Inf--Inf) as input
#> ! [2025-07-26 07:18:39] Error in `object[[graph]]`:
#> ! [2025-07-26 07:18:39] ! ‘Uncorrected_SNN’ not found in this Seurat object
#> ! [2025-07-26 07:18:39]
#> ! [2025-07-26 07:18:39] Error when performing nonlinear dimension reduction. Skip this step...
#> ℹ [2025-07-26 07:18:40] Uncorrected_integrate done
#> ℹ [2025-07-26 07:18:40] Elapsed time: 8.74 secs
CellDimPlot(
panc8_sub,
group.by = c("tech", "celltype")
)
#> Warning: No shared levels found between `names(values)` of the manual scale and the
#> data's fill values.
#> Warning: No shared levels found between `names(values)` of the manual scale and the
#> data's fill values.
panc8_sub <- integration_scop(
panc8_sub,
batch = "tech",
integration_method = "Uncorrected",
HVF_min_intersection = 5,
scale_within_batch = TRUE
)
#> ℹ [2025-07-26 07:18:31] Start Uncorrected_integrate
#> ℹ [2025-07-26 07:18:31] Spliting srt_merge into srt_list by column tech...
#> ℹ [2025-07-26 07:18:33] Checking srt_list...
#> Warning: The following features were labelled as variable in 'var.features' but had no corresponding rank in `var.features.rank` and will therefore be ignored: 'AHNAK', 'ANXA3', 'CDKN1A', 'CFI', 'CLDN1', 'CNN3', 'CTSH', 'DUSP1', 'FAM84A', 'GCNT3', 'GC', 'ID1', 'KLF6', 'LAD1', 'MECOM', 'MSN', 'MTUS1', 'PALLD', 'PHGDH', 'PLK2', 'PPP2R2B', 'RHOC', 'S100A11', 'S100A16', 'SCTR', 'SLC43A3', 'SLC4A4', 'SOX9', 'SPOCK1', 'SYNPO', 'TC2N', 'TFPI2', 'TRIB3', 'VTCN1', 'XBP1', 'ZFP36L1'
#> ℹ [2025-07-26 07:18:33] Data 1/5 of the srt_list has been log-normalized.
#> ℹ [2025-07-26 07:18:33] Perform FindVariableFeatures on the data 1/5 of the srt_list...
#> Warning: The following features were labelled as variable in 'var.features' but had no corresponding rank in `var.features.rank` and will therefore be ignored: 'AHNAK', 'ANXA3', 'CDKN1A', 'CFI', 'CLDN1', 'CNN3', 'CTSH', 'DUSP1', 'FAM84A', 'GCNT3', 'GC', 'ID1', 'KLF6', 'LAD1', 'MECOM', 'MSN', 'MTUS1', 'PALLD', 'PHGDH', 'PLK2', 'PPP2R2B', 'RHOC', 'S100A11', 'S100A16', 'SCTR', 'SLC43A3', 'SLC4A4', 'SOX9', 'SPOCK1', 'SYNPO', 'TC2N', 'TFPI2', 'TRIB3', 'VTCN1', 'XBP1', 'ZFP36L1'
#> ℹ [2025-07-26 07:18:34] Data 2/5 of the srt_list has been log-normalized.
#> ℹ [2025-07-26 07:18:34] Perform FindVariableFeatures on the data 2/5 of the srt_list...
#> Warning: The following features were labelled as variable in 'var.features' but had no corresponding rank in `var.features.rank` and will therefore be ignored: 'AHNAK', 'ANXA3', 'CDKN1A', 'CFI', 'CLDN1', 'CNN3', 'CTSH', 'DUSP1', 'FAM84A', 'GCNT3', 'GC', 'ID1', 'KLF6', 'LAD1', 'MECOM', 'MSN', 'MTUS1', 'PALLD', 'PHGDH', 'PLK2', 'PPP2R2B', 'RHOC', 'S100A11', 'S100A16', 'SCTR', 'SLC43A3', 'SLC4A4', 'SOX9', 'SPOCK1', 'SYNPO', 'TC2N', 'TFPI2', 'TRIB3', 'VTCN1', 'XBP1', 'ZFP36L1'
#> ℹ [2025-07-26 07:18:34] Data 3/5 of the srt_list has been log-normalized.
#> ℹ [2025-07-26 07:18:34] Perform FindVariableFeatures on the data 3/5 of the srt_list...
#> Warning: The following features were labelled as variable in 'var.features' but had no corresponding rank in `var.features.rank` and will therefore be ignored: 'AHNAK', 'ANXA3', 'CDKN1A', 'CFI', 'CLDN1', 'CNN3', 'CTSH', 'DUSP1', 'FAM84A', 'GCNT3', 'GC', 'ID1', 'KLF6', 'LAD1', 'MECOM', 'MSN', 'MTUS1', 'PALLD', 'PHGDH', 'PLK2', 'PPP2R2B', 'RHOC', 'S100A11', 'S100A16', 'SCTR', 'SLC43A3', 'SLC4A4', 'SOX9', 'SPOCK1', 'SYNPO', 'TC2N', 'TFPI2', 'TRIB3', 'VTCN1', 'XBP1', 'ZFP36L1'
#> ℹ [2025-07-26 07:18:35] Data 4/5 of the srt_list has been log-normalized.
#> ℹ [2025-07-26 07:18:35] Perform FindVariableFeatures on the data 4/5 of the srt_list...
#> Warning: The following features were labelled as variable in 'var.features' but had no corresponding rank in `var.features.rank` and will therefore be ignored: 'AHNAK', 'ANXA3', 'CDKN1A', 'CFI', 'CLDN1', 'CNN3', 'CTSH', 'DUSP1', 'FAM84A', 'GCNT3', 'GC', 'ID1', 'KLF6', 'LAD1', 'MECOM', 'MSN', 'MTUS1', 'PALLD', 'PHGDH', 'PLK2', 'PPP2R2B', 'RHOC', 'S100A11', 'S100A16', 'SCTR', 'SLC43A3', 'SLC4A4', 'SOX9', 'SPOCK1', 'SYNPO', 'TC2N', 'TFPI2', 'TRIB3', 'VTCN1', 'XBP1', 'ZFP36L1'
#> ℹ [2025-07-26 07:18:35] Data 5/5 of the srt_list has been log-normalized.
#> ℹ [2025-07-26 07:18:35] Perform FindVariableFeatures on the data 5/5 of the srt_list...
#> ℹ [2025-07-26 07:18:36] Use the separate HVF from srt_list...
#> ℹ [2025-07-26 07:18:36] Number of available HVF: 259
#> ℹ [2025-07-26 07:18:36] Finished checking.
#> ℹ [2025-07-26 07:18:38] Perform integration(Uncorrected) on the data...
#> ℹ [2025-07-26 07:18:38] Perform ScaleData on the data...
#> ℹ [2025-07-26 07:18:38] Perform linear dimension reduction (pca) on the data...
#> ℹ [2025-07-26 07:18:38] Linear_reduction(pca) is already existed. Skip calculation.
#> Warning: The following arguments are not used: features
#> Warning: multiple layers are identified by counts.1.1.1.1 counts.2.1 counts.2.1.1 counts.2.1.1.1 data.1.1.1.1 scale.data.1.1.1.1 data.2.1.1.1 scale.data.2.1.1.1 data.2.1.1 scale.data.2.1.1 data.2.1 scale.data.2.1
#> only the first layer is used
#> ! [2025-07-26 07:18:38] Error: Not compatible with requested type: [type=S4; target=double].
#> ! [2025-07-26 07:18:38] Error when performing FindClusters. Skip this step...
#> ℹ [2025-07-26 07:18:38] Perform nonlinear dimension reduction (umap) on the data...
#> Warning: no non-missing arguments to min; returning Inf
#> Warning: no non-missing arguments to max; returning -Inf
#> ℹ [2025-07-26 07:18:39] Non-linear dimensionality reduction(umap) using Reduction(Uncorrectedpca, dims:Inf--Inf) as input
#> ! [2025-07-26 07:18:39] Error in `object[[graph]]`:
#> ! [2025-07-26 07:18:39] ! ‘Uncorrected_SNN’ not found in this Seurat object
#> ! [2025-07-26 07:18:39]
#> ! [2025-07-26 07:18:39] Error when performing nonlinear dimension reduction. Skip this step...
#> ℹ [2025-07-26 07:18:40] Uncorrected_integrate done
#> ℹ [2025-07-26 07:18:40] Elapsed time: 8.74 secs
CellDimPlot(
panc8_sub,
group.by = c("tech", "celltype")
)
#> Warning: No shared levels found between `names(values)` of the manual scale and the
#> data's fill values.
#> Warning: No shared levels found between `names(values)` of the manual scale and the
#> data's fill values.
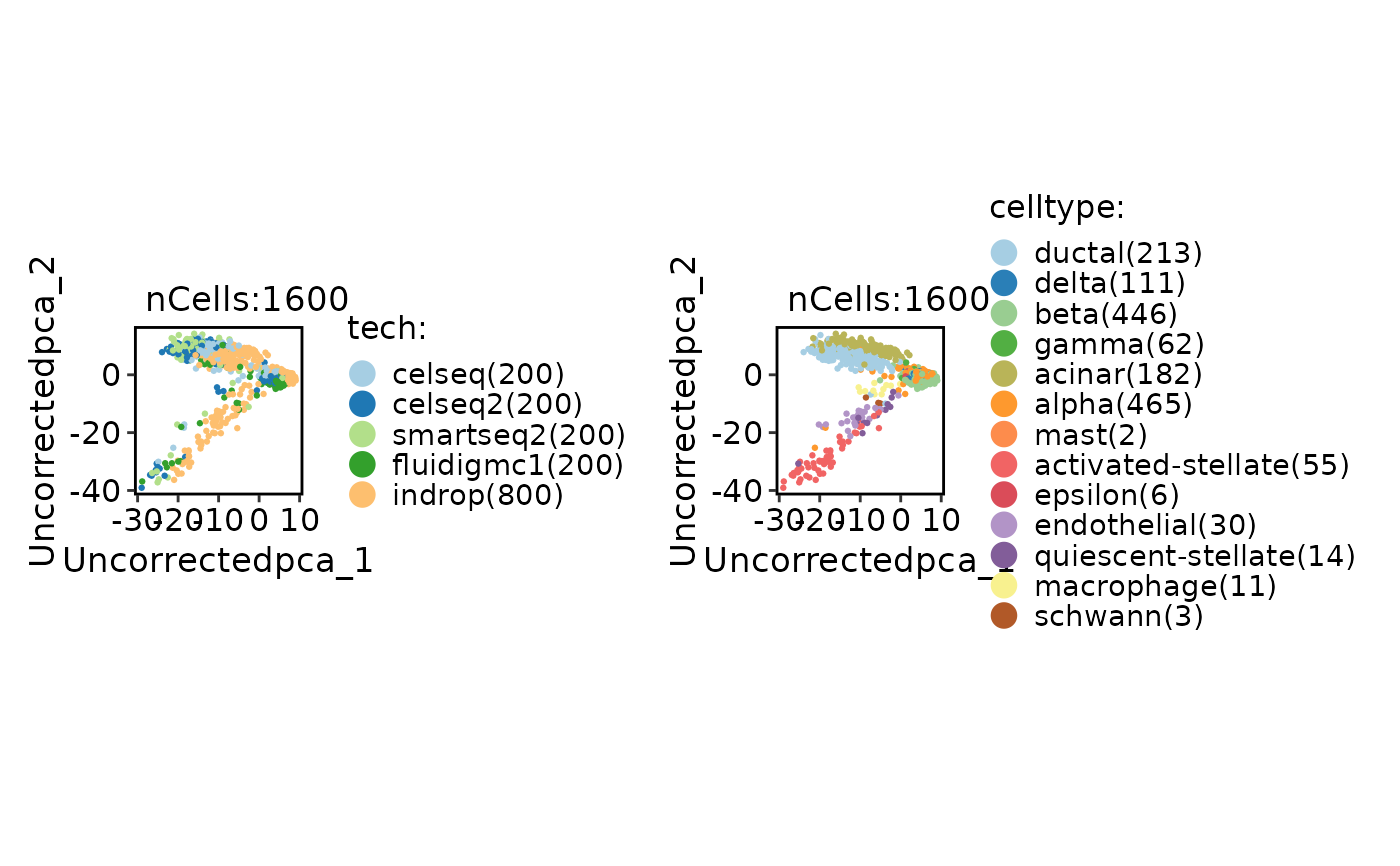 panc8_sub <- integration_scop(
panc8_sub,
batch = "tech",
integration_method = "Seurat"
)
#> ℹ [2025-07-26 07:18:40] Start Seurat_integrate
#> ℹ [2025-07-26 07:18:40] Spliting srt_merge into srt_list by column tech...
#> ℹ [2025-07-26 07:18:42] Checking srt_list...
#> Warning: The following features were labelled as variable in 'var.features' but had no corresponding rank in `var.features.rank` and will therefore be ignored: 'AHNAK', 'ANXA3', 'CDKN1A', 'CFI', 'CLDN1', 'CNN3', 'CTSH', 'DUSP1', 'FAM84A', 'GCNT3', 'GC', 'ID1', 'KLF6', 'LAD1', 'MECOM', 'MSN', 'MTUS1', 'PALLD', 'PHGDH', 'PLK2', 'PPP2R2B', 'RHOC', 'S100A11', 'S100A16', 'SCTR', 'SLC43A3', 'SLC4A4', 'SOX9', 'SPOCK1', 'SYNPO', 'TC2N', 'TFPI2', 'TRIB3', 'VTCN1', 'XBP1', 'ZFP36L1'
#> ℹ [2025-07-26 07:18:42] Data 1/5 of the srt_list has been log-normalized.
#> ℹ [2025-07-26 07:18:42] Perform FindVariableFeatures on the data 1/5 of the srt_list...
#> Warning: The following features were labelled as variable in 'var.features' but had no corresponding rank in `var.features.rank` and will therefore be ignored: 'AHNAK', 'ANXA3', 'CDKN1A', 'CFI', 'CLDN1', 'CNN3', 'CTSH', 'DUSP1', 'FAM84A', 'GCNT3', 'GC', 'ID1', 'KLF6', 'LAD1', 'MECOM', 'MSN', 'MTUS1', 'PALLD', 'PHGDH', 'PLK2', 'PPP2R2B', 'RHOC', 'S100A11', 'S100A16', 'SCTR', 'SLC43A3', 'SLC4A4', 'SOX9', 'SPOCK1', 'SYNPO', 'TC2N', 'TFPI2', 'TRIB3', 'VTCN1', 'XBP1', 'ZFP36L1'
#> ℹ [2025-07-26 07:18:43] Data 2/5 of the srt_list has been log-normalized.
#> ℹ [2025-07-26 07:18:43] Perform FindVariableFeatures on the data 2/5 of the srt_list...
#> Warning: The following features were labelled as variable in 'var.features' but had no corresponding rank in `var.features.rank` and will therefore be ignored: 'AHNAK', 'ANXA3', 'CDKN1A', 'CFI', 'CLDN1', 'CNN3', 'CTSH', 'DUSP1', 'FAM84A', 'GCNT3', 'GC', 'ID1', 'KLF6', 'LAD1', 'MECOM', 'MSN', 'MTUS1', 'PALLD', 'PHGDH', 'PLK2', 'PPP2R2B', 'RHOC', 'S100A11', 'S100A16', 'SCTR', 'SLC43A3', 'SLC4A4', 'SOX9', 'SPOCK1', 'SYNPO', 'TC2N', 'TFPI2', 'TRIB3', 'VTCN1', 'XBP1', 'ZFP36L1'
#> ℹ [2025-07-26 07:18:43] Data 3/5 of the srt_list has been log-normalized.
#> ℹ [2025-07-26 07:18:43] Perform FindVariableFeatures on the data 3/5 of the srt_list...
#> Warning: The following features were labelled as variable in 'var.features' but had no corresponding rank in `var.features.rank` and will therefore be ignored: 'AHNAK', 'ANXA3', 'CDKN1A', 'CFI', 'CLDN1', 'CNN3', 'CTSH', 'DUSP1', 'FAM84A', 'GCNT3', 'GC', 'ID1', 'KLF6', 'LAD1', 'MECOM', 'MSN', 'MTUS1', 'PALLD', 'PHGDH', 'PLK2', 'PPP2R2B', 'RHOC', 'S100A11', 'S100A16', 'SCTR', 'SLC43A3', 'SLC4A4', 'SOX9', 'SPOCK1', 'SYNPO', 'TC2N', 'TFPI2', 'TRIB3', 'VTCN1', 'XBP1', 'ZFP36L1'
#> ℹ [2025-07-26 07:18:44] Data 4/5 of the srt_list has been log-normalized.
#> ℹ [2025-07-26 07:18:44] Perform FindVariableFeatures on the data 4/5 of the srt_list...
#> Warning: The following features were labelled as variable in 'var.features' but had no corresponding rank in `var.features.rank` and will therefore be ignored: 'AHNAK', 'ANXA3', 'CDKN1A', 'CFI', 'CLDN1', 'CNN3', 'CTSH', 'DUSP1', 'FAM84A', 'GCNT3', 'GC', 'ID1', 'KLF6', 'LAD1', 'MECOM', 'MSN', 'MTUS1', 'PALLD', 'PHGDH', 'PLK2', 'PPP2R2B', 'RHOC', 'S100A11', 'S100A16', 'SCTR', 'SLC43A3', 'SLC4A4', 'SOX9', 'SPOCK1', 'SYNPO', 'TC2N', 'TFPI2', 'TRIB3', 'VTCN1', 'XBP1', 'ZFP36L1'
#> ℹ [2025-07-26 07:18:44] Data 5/5 of the srt_list has been log-normalized.
#> ℹ [2025-07-26 07:18:44] Perform FindVariableFeatures on the data 5/5 of the srt_list...
#> ℹ [2025-07-26 07:18:45] Use the separate HVF from srt_list...
#> ℹ [2025-07-26 07:18:45] Number of available HVF: 2000
#> ℹ [2025-07-26 07:18:45] Finished checking.
#> ℹ [2025-07-26 07:18:47] Perform FindIntegrationAnchors on the data...
#> Warning: Different features in new layer data than already exists for scale.data
#> Warning: Different features in new layer data than already exists for scale.data
#> Warning: Different features in new layer data than already exists for scale.data
#> Warning: Different features in new layer data than already exists for scale.data
#> Warning: Different features in new layer data than already exists for scale.data
#> Warning: The `slot` argument of `GetAssayData()` is deprecated as of SeuratObject 5.0.0.
#> ℹ Please use the `layer` argument instead.
#> ℹ The deprecated feature was likely used in the Seurat package.
#> Please report the issue at <https://github.com/satijalab/seurat/issues>.
#> Warning: The `slot` argument of `SetAssayData()` is deprecated as of SeuratObject 5.0.0.
#> ℹ Please use the `layer` argument instead.
#> ℹ The deprecated feature was likely used in the Seurat package.
#> Please report the issue at <https://github.com/satijalab/seurat/issues>.
#> ℹ [2025-07-26 07:19:18] Perform integration(Seurat) on the data...
#> Warning: Layer counts isn't present in the assay object; returning NULL
#> Warning: Layer counts isn't present in the assay object; returning NULL
#> Warning: Layer counts isn't present in the assay object; returning NULL
#> Warning: Different cells in new layer data than already exists for scale.data
#> Warning: Layer counts isn't present in the assay object; returning NULL
#> ℹ [2025-07-26 07:19:30] Perform ScaleData on the data...
#> ℹ [2025-07-26 07:19:30] Perform linear dimension reduction (pca) on the data...
#> ℹ [2025-07-26 07:19:31] Perform FindClusters (louvain) on the data...
#> ℹ [2025-07-26 07:19:31] Reorder clusters...
#> ℹ [2025-07-26 07:19:31] Using 'Seurat::AverageExpression()' to calculate pseudo-bulk data for 'Assay'.
#> ℹ [2025-07-26 07:19:31] Perform nonlinear dimension reduction (umap) on the data...
#> ℹ [2025-07-26 07:19:31] Non-linear dimensionality reduction(umap) using Reduction(Seuratpca, dims:1-12) as input
#> ℹ [2025-07-26 07:19:35] Non-linear dimensionality reduction(umap) using Reduction(Seuratpca, dims:1-12) as input
#> ℹ [2025-07-26 07:19:40] Seurat_integrate done
#> ℹ [2025-07-26 07:19:40] Elapsed time: 59.61 secs
CellDimPlot(panc8_sub, group.by = c("tech", "celltype"))
#> Warning: No shared levels found between `names(values)` of the manual scale and the
#> data's fill values.
#> Warning: No shared levels found between `names(values)` of the manual scale and the
#> data's fill values.
panc8_sub <- integration_scop(
panc8_sub,
batch = "tech",
integration_method = "Seurat"
)
#> ℹ [2025-07-26 07:18:40] Start Seurat_integrate
#> ℹ [2025-07-26 07:18:40] Spliting srt_merge into srt_list by column tech...
#> ℹ [2025-07-26 07:18:42] Checking srt_list...
#> Warning: The following features were labelled as variable in 'var.features' but had no corresponding rank in `var.features.rank` and will therefore be ignored: 'AHNAK', 'ANXA3', 'CDKN1A', 'CFI', 'CLDN1', 'CNN3', 'CTSH', 'DUSP1', 'FAM84A', 'GCNT3', 'GC', 'ID1', 'KLF6', 'LAD1', 'MECOM', 'MSN', 'MTUS1', 'PALLD', 'PHGDH', 'PLK2', 'PPP2R2B', 'RHOC', 'S100A11', 'S100A16', 'SCTR', 'SLC43A3', 'SLC4A4', 'SOX9', 'SPOCK1', 'SYNPO', 'TC2N', 'TFPI2', 'TRIB3', 'VTCN1', 'XBP1', 'ZFP36L1'
#> ℹ [2025-07-26 07:18:42] Data 1/5 of the srt_list has been log-normalized.
#> ℹ [2025-07-26 07:18:42] Perform FindVariableFeatures on the data 1/5 of the srt_list...
#> Warning: The following features were labelled as variable in 'var.features' but had no corresponding rank in `var.features.rank` and will therefore be ignored: 'AHNAK', 'ANXA3', 'CDKN1A', 'CFI', 'CLDN1', 'CNN3', 'CTSH', 'DUSP1', 'FAM84A', 'GCNT3', 'GC', 'ID1', 'KLF6', 'LAD1', 'MECOM', 'MSN', 'MTUS1', 'PALLD', 'PHGDH', 'PLK2', 'PPP2R2B', 'RHOC', 'S100A11', 'S100A16', 'SCTR', 'SLC43A3', 'SLC4A4', 'SOX9', 'SPOCK1', 'SYNPO', 'TC2N', 'TFPI2', 'TRIB3', 'VTCN1', 'XBP1', 'ZFP36L1'
#> ℹ [2025-07-26 07:18:43] Data 2/5 of the srt_list has been log-normalized.
#> ℹ [2025-07-26 07:18:43] Perform FindVariableFeatures on the data 2/5 of the srt_list...
#> Warning: The following features were labelled as variable in 'var.features' but had no corresponding rank in `var.features.rank` and will therefore be ignored: 'AHNAK', 'ANXA3', 'CDKN1A', 'CFI', 'CLDN1', 'CNN3', 'CTSH', 'DUSP1', 'FAM84A', 'GCNT3', 'GC', 'ID1', 'KLF6', 'LAD1', 'MECOM', 'MSN', 'MTUS1', 'PALLD', 'PHGDH', 'PLK2', 'PPP2R2B', 'RHOC', 'S100A11', 'S100A16', 'SCTR', 'SLC43A3', 'SLC4A4', 'SOX9', 'SPOCK1', 'SYNPO', 'TC2N', 'TFPI2', 'TRIB3', 'VTCN1', 'XBP1', 'ZFP36L1'
#> ℹ [2025-07-26 07:18:43] Data 3/5 of the srt_list has been log-normalized.
#> ℹ [2025-07-26 07:18:43] Perform FindVariableFeatures on the data 3/5 of the srt_list...
#> Warning: The following features were labelled as variable in 'var.features' but had no corresponding rank in `var.features.rank` and will therefore be ignored: 'AHNAK', 'ANXA3', 'CDKN1A', 'CFI', 'CLDN1', 'CNN3', 'CTSH', 'DUSP1', 'FAM84A', 'GCNT3', 'GC', 'ID1', 'KLF6', 'LAD1', 'MECOM', 'MSN', 'MTUS1', 'PALLD', 'PHGDH', 'PLK2', 'PPP2R2B', 'RHOC', 'S100A11', 'S100A16', 'SCTR', 'SLC43A3', 'SLC4A4', 'SOX9', 'SPOCK1', 'SYNPO', 'TC2N', 'TFPI2', 'TRIB3', 'VTCN1', 'XBP1', 'ZFP36L1'
#> ℹ [2025-07-26 07:18:44] Data 4/5 of the srt_list has been log-normalized.
#> ℹ [2025-07-26 07:18:44] Perform FindVariableFeatures on the data 4/5 of the srt_list...
#> Warning: The following features were labelled as variable in 'var.features' but had no corresponding rank in `var.features.rank` and will therefore be ignored: 'AHNAK', 'ANXA3', 'CDKN1A', 'CFI', 'CLDN1', 'CNN3', 'CTSH', 'DUSP1', 'FAM84A', 'GCNT3', 'GC', 'ID1', 'KLF6', 'LAD1', 'MECOM', 'MSN', 'MTUS1', 'PALLD', 'PHGDH', 'PLK2', 'PPP2R2B', 'RHOC', 'S100A11', 'S100A16', 'SCTR', 'SLC43A3', 'SLC4A4', 'SOX9', 'SPOCK1', 'SYNPO', 'TC2N', 'TFPI2', 'TRIB3', 'VTCN1', 'XBP1', 'ZFP36L1'
#> ℹ [2025-07-26 07:18:44] Data 5/5 of the srt_list has been log-normalized.
#> ℹ [2025-07-26 07:18:44] Perform FindVariableFeatures on the data 5/5 of the srt_list...
#> ℹ [2025-07-26 07:18:45] Use the separate HVF from srt_list...
#> ℹ [2025-07-26 07:18:45] Number of available HVF: 2000
#> ℹ [2025-07-26 07:18:45] Finished checking.
#> ℹ [2025-07-26 07:18:47] Perform FindIntegrationAnchors on the data...
#> Warning: Different features in new layer data than already exists for scale.data
#> Warning: Different features in new layer data than already exists for scale.data
#> Warning: Different features in new layer data than already exists for scale.data
#> Warning: Different features in new layer data than already exists for scale.data
#> Warning: Different features in new layer data than already exists for scale.data
#> Warning: The `slot` argument of `GetAssayData()` is deprecated as of SeuratObject 5.0.0.
#> ℹ Please use the `layer` argument instead.
#> ℹ The deprecated feature was likely used in the Seurat package.
#> Please report the issue at <https://github.com/satijalab/seurat/issues>.
#> Warning: The `slot` argument of `SetAssayData()` is deprecated as of SeuratObject 5.0.0.
#> ℹ Please use the `layer` argument instead.
#> ℹ The deprecated feature was likely used in the Seurat package.
#> Please report the issue at <https://github.com/satijalab/seurat/issues>.
#> ℹ [2025-07-26 07:19:18] Perform integration(Seurat) on the data...
#> Warning: Layer counts isn't present in the assay object; returning NULL
#> Warning: Layer counts isn't present in the assay object; returning NULL
#> Warning: Layer counts isn't present in the assay object; returning NULL
#> Warning: Different cells in new layer data than already exists for scale.data
#> Warning: Layer counts isn't present in the assay object; returning NULL
#> ℹ [2025-07-26 07:19:30] Perform ScaleData on the data...
#> ℹ [2025-07-26 07:19:30] Perform linear dimension reduction (pca) on the data...
#> ℹ [2025-07-26 07:19:31] Perform FindClusters (louvain) on the data...
#> ℹ [2025-07-26 07:19:31] Reorder clusters...
#> ℹ [2025-07-26 07:19:31] Using 'Seurat::AverageExpression()' to calculate pseudo-bulk data for 'Assay'.
#> ℹ [2025-07-26 07:19:31] Perform nonlinear dimension reduction (umap) on the data...
#> ℹ [2025-07-26 07:19:31] Non-linear dimensionality reduction(umap) using Reduction(Seuratpca, dims:1-12) as input
#> ℹ [2025-07-26 07:19:35] Non-linear dimensionality reduction(umap) using Reduction(Seuratpca, dims:1-12) as input
#> ℹ [2025-07-26 07:19:40] Seurat_integrate done
#> ℹ [2025-07-26 07:19:40] Elapsed time: 59.61 secs
CellDimPlot(panc8_sub, group.by = c("tech", "celltype"))
#> Warning: No shared levels found between `names(values)` of the manual scale and the
#> data's fill values.
#> Warning: No shared levels found between `names(values)` of the manual scale and the
#> data's fill values.
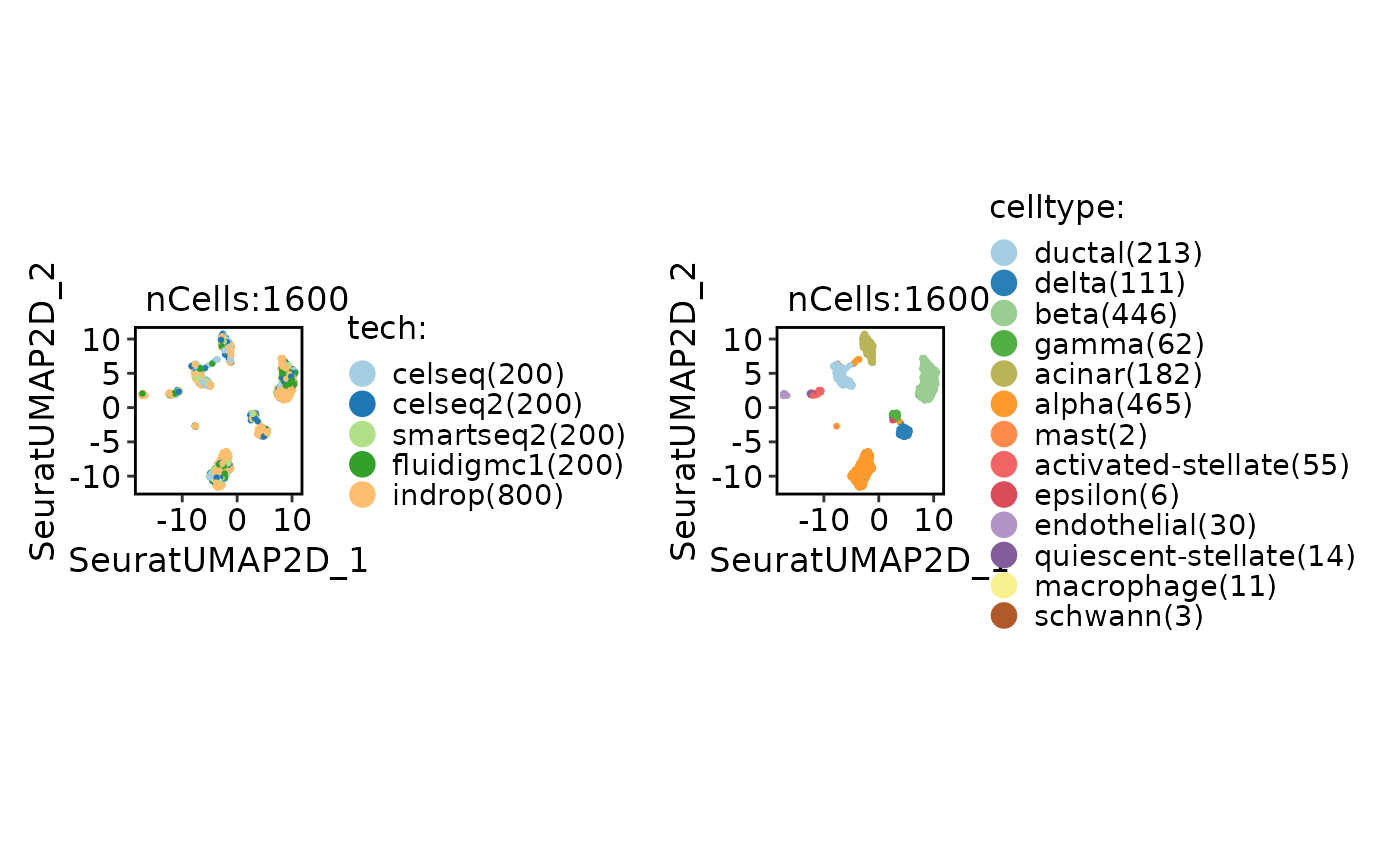 if (FALSE) { # \dontrun{
panc8_sub <- integration_scop(
panc8_sub,
batch = "tech",
integration_method = "Seurat",
FindIntegrationAnchors_params = list(reduction = "rpca")
)
CellDimPlot(panc8_sub, group.by = c("tech", "celltype"))
integration_methods <- c(
"Uncorrected", "Seurat", "scVI", "MNN", "fastMNN", "Harmony",
"Scanorama", "BBKNN", "CSS", "LIGER", "Conos", "ComBat"
)
for (method in integration_methods) {
panc8_sub <- integration_scop(
panc8_sub,
batch = "tech",
integration_method = method,
linear_reduction_dims_use = 1:50,
nonlinear_reduction = "umap"
)
print(
CellDimPlot(panc8_sub,
group.by = c("tech", "celltype"),
reduction = paste0(method, "UMAP2D"),
xlab = "", ylab = "", title = method,
legend.position = "none", theme_use = "theme_blank"
)
)
}
nonlinear_reductions <- c(
"umap", "tsne", "dm", "phate",
"pacmap", "trimap", "largevis", "fr"
)
panc8_sub <- integration_scop(
panc8_sub,
batch = "tech",
integration_method = "Seurat",
linear_reduction_dims_use = 1:50,
nonlinear_reduction = nonlinear_reductions
)
for (nr in nonlinear_reductions) {
print(
CellDimPlot(
panc8_sub,
group.by = c("tech", "celltype"),
reduction = paste0("Seurat", nr, "2D"),
xlab = "", ylab = "", title = nr,
legend.position = "none", theme_use = "theme_blank"
)
)
}
} # }
if (FALSE) { # \dontrun{
panc8_sub <- integration_scop(
panc8_sub,
batch = "tech",
integration_method = "Seurat",
FindIntegrationAnchors_params = list(reduction = "rpca")
)
CellDimPlot(panc8_sub, group.by = c("tech", "celltype"))
integration_methods <- c(
"Uncorrected", "Seurat", "scVI", "MNN", "fastMNN", "Harmony",
"Scanorama", "BBKNN", "CSS", "LIGER", "Conos", "ComBat"
)
for (method in integration_methods) {
panc8_sub <- integration_scop(
panc8_sub,
batch = "tech",
integration_method = method,
linear_reduction_dims_use = 1:50,
nonlinear_reduction = "umap"
)
print(
CellDimPlot(panc8_sub,
group.by = c("tech", "celltype"),
reduction = paste0(method, "UMAP2D"),
xlab = "", ylab = "", title = method,
legend.position = "none", theme_use = "theme_blank"
)
)
}
nonlinear_reductions <- c(
"umap", "tsne", "dm", "phate",
"pacmap", "trimap", "largevis", "fr"
)
panc8_sub <- integration_scop(
panc8_sub,
batch = "tech",
integration_method = "Seurat",
linear_reduction_dims_use = 1:50,
nonlinear_reduction = nonlinear_reductions
)
for (nr in nonlinear_reductions) {
print(
CellDimPlot(
panc8_sub,
group.by = c("tech", "celltype"),
reduction = paste0("Seurat", nr, "2D"),
xlab = "", ylab = "", title = nr,
legend.position = "none", theme_use = "theme_blank"
)
)
}
} # }