Run doublet-calling with scds
Arguments
- srt
A Seurat object.
- assay
The name of the assay to be used for doublet-calling. Default is
"RNA".- db_rate
The expected doublet rate. Default is calculated as
ncol(srt) / 1000 * 0.01.- method
The method to be used for doublet-calling. Options are
"hybrid","cxds", or"bcds".- ...
Additional arguments to be passed to
scds::cxds_bcds_hybrid().
Examples
data(pancreas_sub)
pancreas_sub <- standard_scop(pancreas_sub)
#> ℹ [2026-02-11 04:21:03] Start standard scop workflow...
#> ℹ [2026-02-11 04:21:04] Checking a list of <Seurat>...
#> ! [2026-02-11 04:21:04] Data 1/1 of the `srt_list` is "unknown"
#> ℹ [2026-02-11 04:21:04] Perform `NormalizeData()` with `normalization.method = 'LogNormalize'` on the data 1/1 of the `srt_list`...
#> ℹ [2026-02-11 04:21:06] Perform `Seurat::FindVariableFeatures()` on the data 1/1 of the `srt_list`...
#> ℹ [2026-02-11 04:21:06] Use the separate HVF from srt_list
#> ℹ [2026-02-11 04:21:07] Number of available HVF: 2000
#> ℹ [2026-02-11 04:21:07] Finished check
#> ℹ [2026-02-11 04:21:07] Perform `Seurat::ScaleData()`
#> ℹ [2026-02-11 04:21:07] Perform pca linear dimension reduction
#> ℹ [2026-02-11 04:21:08] Perform `Seurat::FindClusters()` with `cluster_algorithm = 'louvain'` and `cluster_resolution = 0.6`
#> ℹ [2026-02-11 04:21:08] Reorder clusters...
#> ℹ [2026-02-11 04:21:08] Perform umap nonlinear dimension reduction
#> ℹ [2026-02-11 04:21:08] Non-linear dimensionality reduction (umap) using (Standardpca) dims (1-50) as input
#> ℹ [2026-02-11 04:21:13] Non-linear dimensionality reduction (umap) using (Standardpca) dims (1-50) as input
#> ✔ [2026-02-11 04:21:17] Run scop standard workflow completed
pancreas_sub <- db_scds(pancreas_sub, method = "hybrid")
#> ℹ [2026-02-11 04:21:18] Data type is raw counts
#> Registered S3 method overwritten by 'pROC':
#> method from
#> plot.roc spatstat.explore
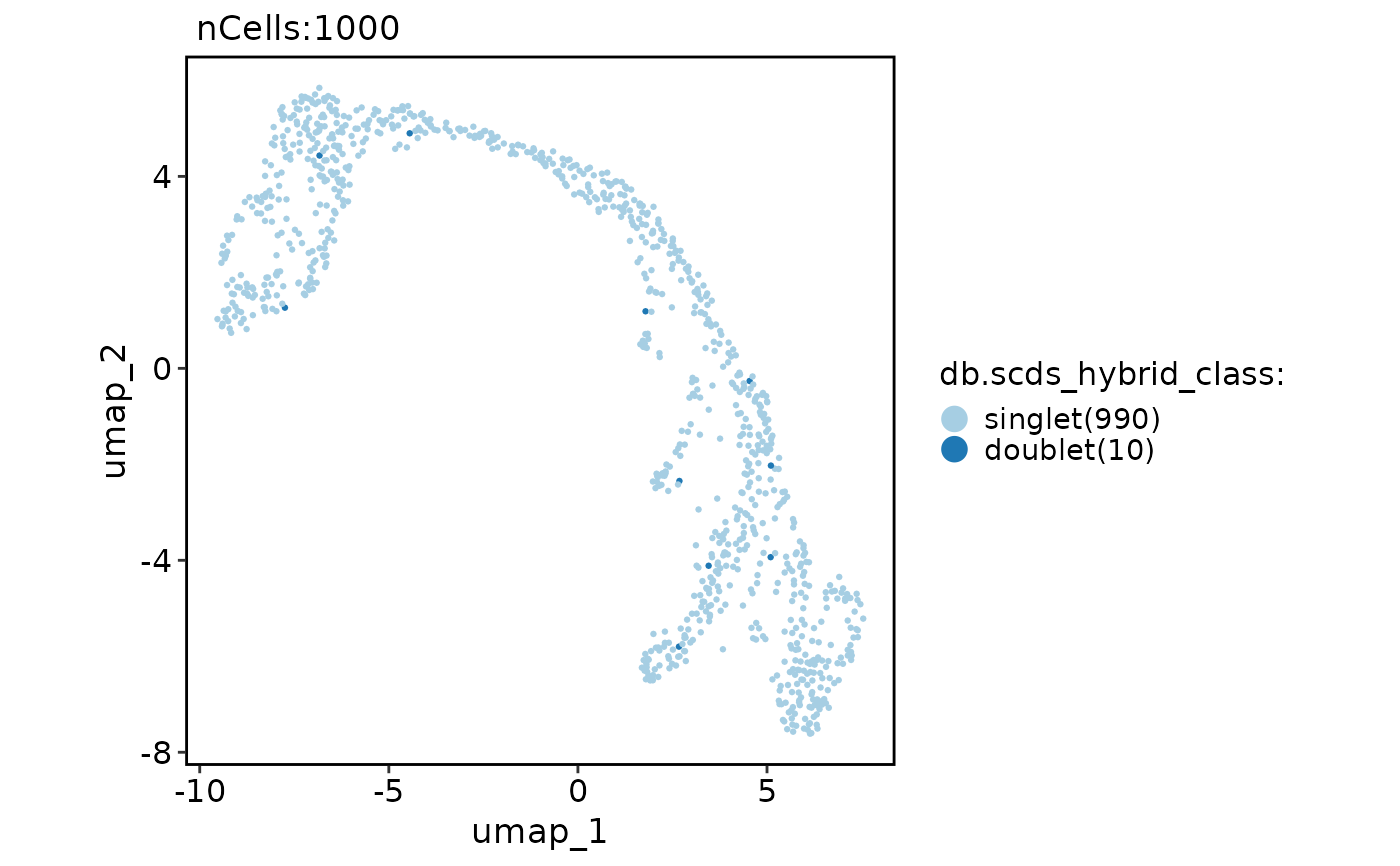
CellDimPlot(
pancreas_sub,
reduction = "umap",
group.by = "db.scds_hybrid_class"
)
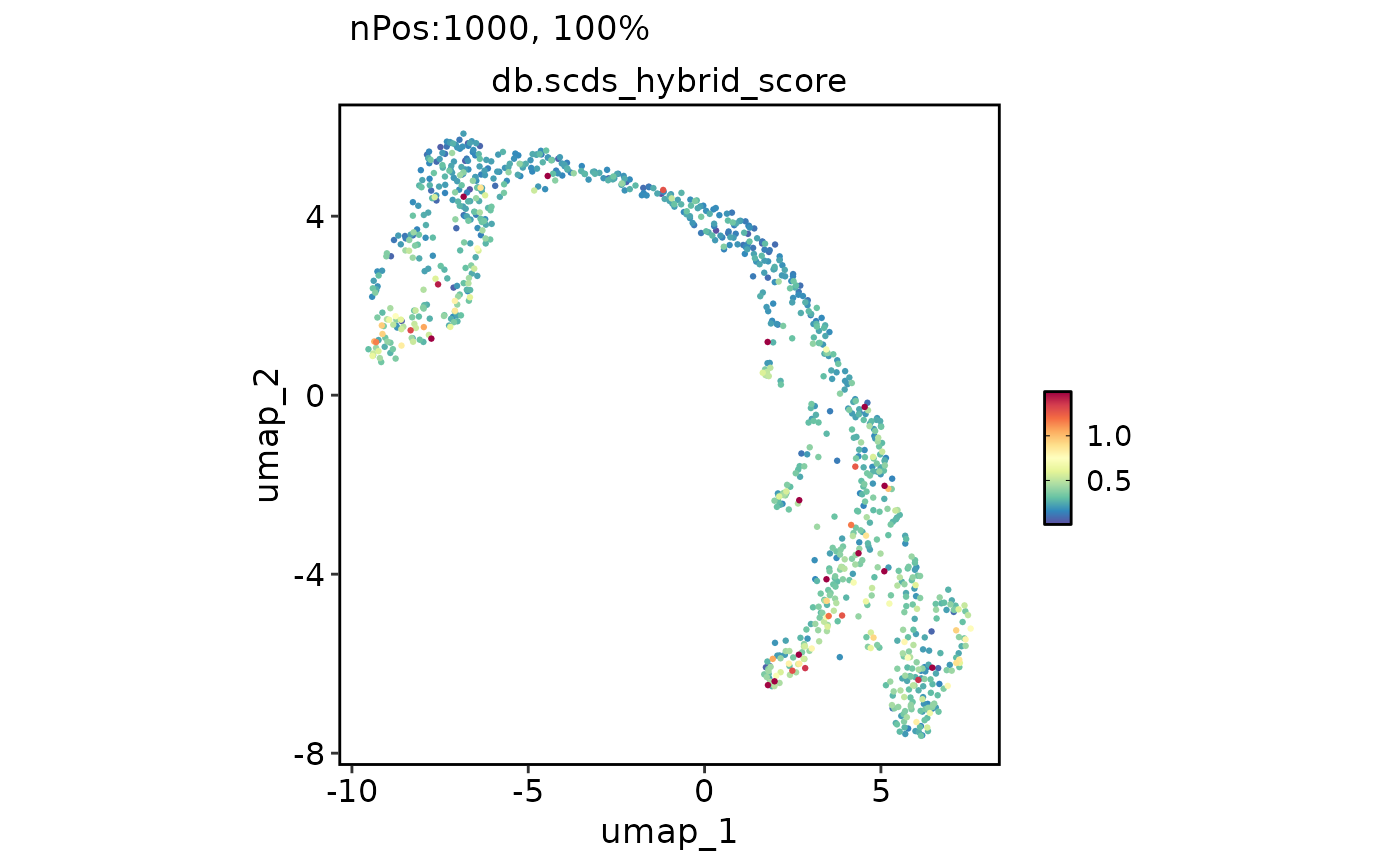
 FeatureDimPlot(
pancreas_sub,
reduction = "umap",
features = "db.scds_hybrid_score"
)
FeatureDimPlot(
pancreas_sub,
reduction = "umap",
features = "db.scds_hybrid_score"
)