Annotate single cells using scmap.
Usage
RunScmap(
srt_query,
srt_ref,
ref_group = NULL,
query_assay = "RNA",
ref_assay = "RNA",
method = "scmapCluster",
nfeatures = 500,
threshold = 0.5,
k = 10
)Arguments
- srt_query
An object of class Seurat to be annotated with cell types.
- srt_ref
An object of class Seurat storing the reference cells.
- ref_group
A character vector specifying the column name in the
srt_refmetadata that represents the cell grouping.- query_assay
A character vector specifying the assay to be used for the query data. Default is the default assay of the
srt_queryobject.- ref_assay
A character vector specifying the assay to be used for the reference data. Default is the default assay of the
srt_refobject.- method
The method to be used for scmap analysis. Can be any of
"scmapCluster"or"scmapCell". Default is"scmapCluster".- nfeatures
The number of top features to be selected. Default is
500.- threshold
The threshold value on similarity to determine if a cell is assigned to a cluster. This should be a value between
0and1. Default is0.5.- k
Number of clusters per group for k-means clustering when
methodis"scmapCell". Default is10.
Examples
data(panc8_sub)
panc8_sub <- standard_scop(panc8_sub)
#> ℹ [2026-02-11 04:08:22] Start standard scop workflow...
#> ℹ [2026-02-11 04:08:23] Checking a list of <Seurat>...
#> ! [2026-02-11 04:08:23] Data 1/1 of the `srt_list` is "unknown"
#> ℹ [2026-02-11 04:08:23] Perform `NormalizeData()` with `normalization.method = 'LogNormalize'` on the data 1/1 of the `srt_list`...
#> ℹ [2026-02-11 04:08:25] Perform `Seurat::FindVariableFeatures()` on the data 1/1 of the `srt_list`...
#> ℹ [2026-02-11 04:08:26] Use the separate HVF from srt_list
#> ℹ [2026-02-11 04:08:26] Number of available HVF: 2000
#> ℹ [2026-02-11 04:08:26] Finished check
#> ℹ [2026-02-11 04:08:26] Perform `Seurat::ScaleData()`
#> ℹ [2026-02-11 04:08:27] Perform pca linear dimension reduction
#> ℹ [2026-02-11 04:08:28] Perform `Seurat::FindClusters()` with `cluster_algorithm = 'louvain'` and `cluster_resolution = 0.6`
#> ℹ [2026-02-11 04:08:28] Reorder clusters...
#> ℹ [2026-02-11 04:08:28] Perform umap nonlinear dimension reduction
#> ℹ [2026-02-11 04:08:28] Non-linear dimensionality reduction (umap) using (Standardpca) dims (1-50) as input
#> ℹ [2026-02-11 04:08:33] Non-linear dimensionality reduction (umap) using (Standardpca) dims (1-50) as input
#> ✔ [2026-02-11 04:08:38] Run scop standard workflow completed
genenames <- make.unique(
thisutils::capitalize(
rownames(panc8_sub),
force_tolower = TRUE
)
)
names(genenames) <- rownames(panc8_sub)
panc8_sub <- RenameFeatures(
panc8_sub,
newnames = genenames
)
#> ℹ [2026-02-11 04:08:38] Rename features for the assay: RNA
panc8_sub <- CheckDataMerge(
panc8_sub,
batch = "tech"
)[["srt_merge"]]
#> ℹ [2026-02-11 04:08:38] Spliting `srt_merge` into `srt_list` by column "tech"...
#> ℹ [2026-02-11 04:08:39] Checking a list of <Seurat>...
#> ℹ [2026-02-11 04:08:40] Data 1/5 of the `srt_list` has been log-normalized
#> ℹ [2026-02-11 04:08:40] Perform `Seurat::FindVariableFeatures()` on the data 1/5 of the `srt_list`...
#> ℹ [2026-02-11 04:08:40] Data 2/5 of the `srt_list` has been log-normalized
#> ℹ [2026-02-11 04:08:40] Perform `Seurat::FindVariableFeatures()` on the data 2/5 of the `srt_list`...
#> ℹ [2026-02-11 04:08:41] Data 3/5 of the `srt_list` has been log-normalized
#> ℹ [2026-02-11 04:08:41] Perform `Seurat::FindVariableFeatures()` on the data 3/5 of the `srt_list`...
#> ℹ [2026-02-11 04:08:41] Data 4/5 of the `srt_list` has been log-normalized
#> ℹ [2026-02-11 04:08:41] Perform `Seurat::FindVariableFeatures()` on the data 4/5 of the `srt_list`...
#> ℹ [2026-02-11 04:08:42] Data 5/5 of the `srt_list` has been log-normalized
#> ℹ [2026-02-11 04:08:42] Perform `Seurat::FindVariableFeatures()` on the data 5/5 of the `srt_list`...
#> ℹ [2026-02-11 04:08:42] Use the separate HVF from srt_list
#> ℹ [2026-02-11 04:08:42] Number of available HVF: 2000
#> ℹ [2026-02-11 04:08:43] Finished check
data(pancreas_sub)
pancreas_sub <- standard_scop(pancreas_sub)
#> ℹ [2026-02-11 04:08:45] Start standard scop workflow...
#> ℹ [2026-02-11 04:08:46] Checking a list of <Seurat>...
#> ! [2026-02-11 04:08:46] Data 1/1 of the `srt_list` is "unknown"
#> ℹ [2026-02-11 04:08:46] Perform `NormalizeData()` with `normalization.method = 'LogNormalize'` on the data 1/1 of the `srt_list`...
#> ℹ [2026-02-11 04:08:48] Perform `Seurat::FindVariableFeatures()` on the data 1/1 of the `srt_list`...
#> ℹ [2026-02-11 04:08:49] Use the separate HVF from srt_list
#> ℹ [2026-02-11 04:08:49] Number of available HVF: 2000
#> ℹ [2026-02-11 04:08:49] Finished check
#> ℹ [2026-02-11 04:08:49] Perform `Seurat::ScaleData()`
#> ℹ [2026-02-11 04:08:49] Perform pca linear dimension reduction
#> ℹ [2026-02-11 04:08:50] Perform `Seurat::FindClusters()` with `cluster_algorithm = 'louvain'` and `cluster_resolution = 0.6`
#> ℹ [2026-02-11 04:08:50] Reorder clusters...
#> ℹ [2026-02-11 04:08:50] Perform umap nonlinear dimension reduction
#> ℹ [2026-02-11 04:08:50] Non-linear dimensionality reduction (umap) using (Standardpca) dims (1-50) as input
#> ℹ [2026-02-11 04:08:55] Non-linear dimensionality reduction (umap) using (Standardpca) dims (1-50) as input
#> ✔ [2026-02-11 04:09:00] Run scop standard workflow completed
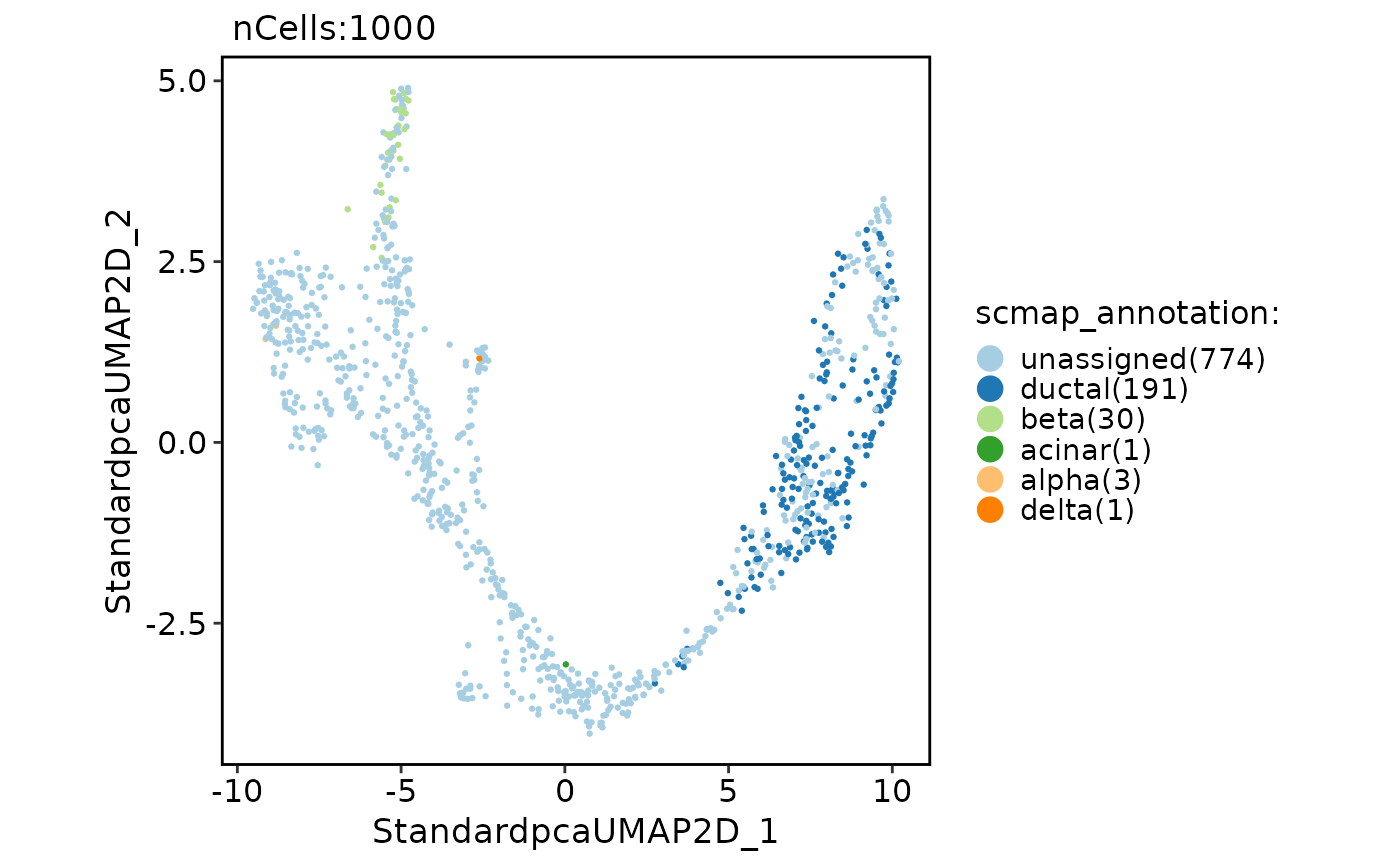
pancreas_sub <- RunScmap(
srt_query = pancreas_sub,
srt_ref = panc8_sub,
ref_group = "celltype",
method = "scmapCluster"
)
#> ℹ [2026-02-11 04:09:28] Data type is log-normalized
#> ℹ [2026-02-11 04:09:28] Detected `srt_query` data type: "log_normalized_counts"
#> ℹ [2026-02-11 04:09:30] Data type is log-normalized
#> ℹ [2026-02-11 04:09:30] Detected `srt_ref` data type: "log_normalized_counts"
#> ℹ [2026-02-11 04:09:32] Perform selectFeatures
#> ℹ [2026-02-11 04:09:32] Perform indexCluster
#> ℹ [2026-02-11 04:09:32] Perform scmapCluster
CellDimPlot(
pancreas_sub,
group.by = "scmap_annotation"
)
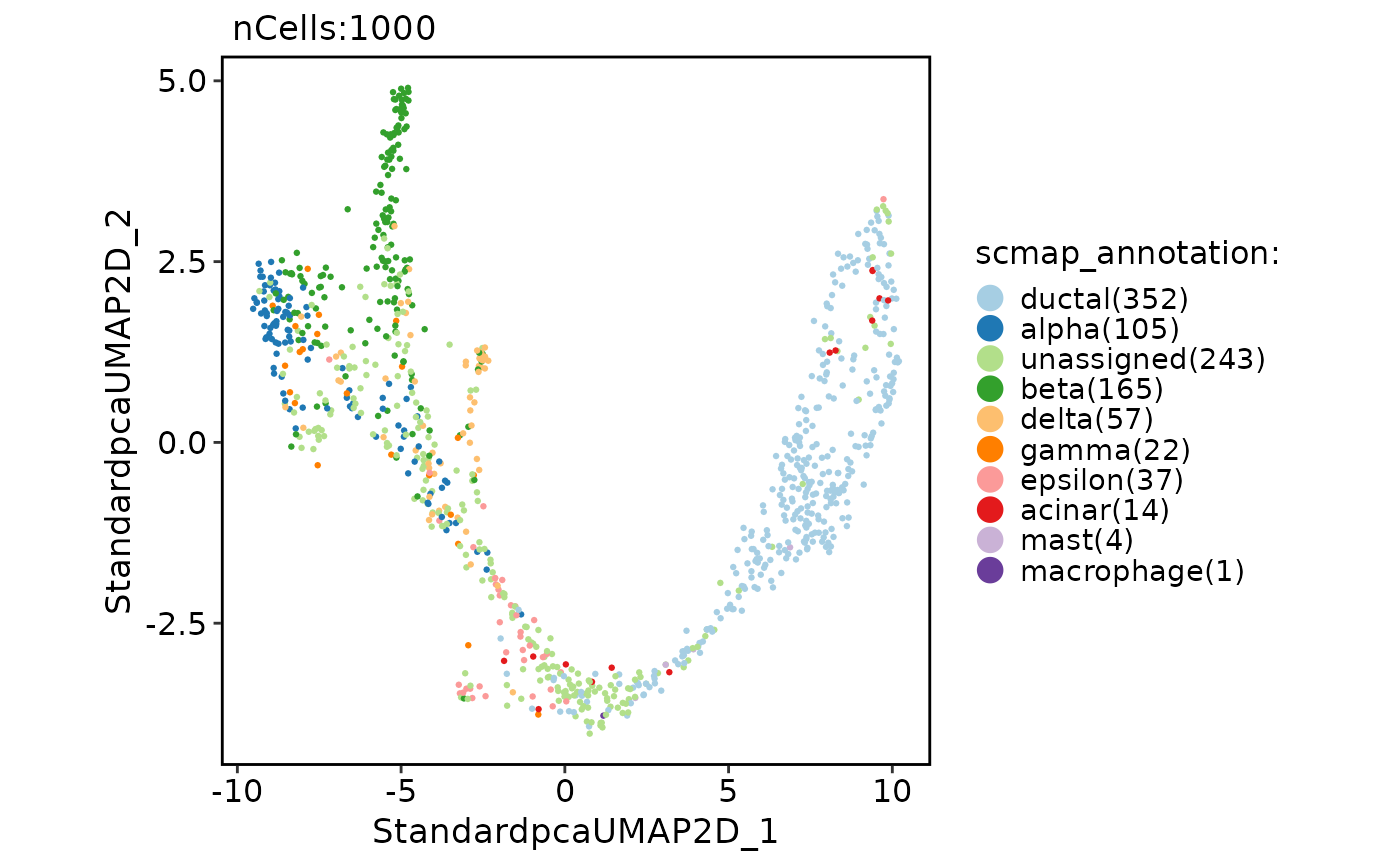 pancreas_sub <- RunScmap(
srt_query = pancreas_sub,
srt_ref = panc8_sub,
ref_group = "celltype",
method = "scmapCell"
)
#> ℹ [2026-02-11 04:09:33] Data type is log-normalized
#> ℹ [2026-02-11 04:09:33] Detected `srt_query` data type: "log_normalized_counts"
#> ℹ [2026-02-11 04:09:36] Data type is log-normalized
#> ℹ [2026-02-11 04:09:36] Detected `srt_ref` data type: "log_normalized_counts"
#> ℹ [2026-02-11 04:09:38] Perform selectFeatures
#> ℹ [2026-02-11 04:09:39] Perform indexCell
#> ℹ [2026-02-11 04:09:39] Perform scmapCell
#> ℹ [2026-02-11 04:09:40] Perform scmapCell2Cluster
CellDimPlot(
pancreas_sub,
group.by = "scmap_annotation"
)
pancreas_sub <- RunScmap(
srt_query = pancreas_sub,
srt_ref = panc8_sub,
ref_group = "celltype",
method = "scmapCell"
)
#> ℹ [2026-02-11 04:09:33] Data type is log-normalized
#> ℹ [2026-02-11 04:09:33] Detected `srt_query` data type: "log_normalized_counts"
#> ℹ [2026-02-11 04:09:36] Data type is log-normalized
#> ℹ [2026-02-11 04:09:36] Detected `srt_ref` data type: "log_normalized_counts"
#> ℹ [2026-02-11 04:09:38] Perform selectFeatures
#> ℹ [2026-02-11 04:09:39] Perform indexCell
#> ℹ [2026-02-11 04:09:39] Perform scmapCell
#> ℹ [2026-02-11 04:09:40] Perform scmapCell2Cluster
CellDimPlot(
pancreas_sub,
group.by = "scmap_annotation"
)