Feature Heatmap
Usage
FeatureHeatmap(
srt,
features = NULL,
cells = NULL,
group.by = NULL,
split.by = NULL,
within_groups = FALSE,
max_cells = 100,
cell_order = NULL,
border = TRUE,
flip = FALSE,
layer = "counts",
assay = NULL,
exp_method = c("zscore", "raw", "fc", "log2fc", "log1p"),
exp_legend_title = NULL,
limits = NULL,
lib_normalize = identical(layer, "counts"),
libsize = NULL,
feature_split = NULL,
feature_split_by = NULL,
n_split = NULL,
split_order = NULL,
split_method = c("kmeans", "hclust", "mfuzz"),
decreasing = FALSE,
fuzzification = NULL,
cluster_features_by = NULL,
cluster_rows = FALSE,
cluster_columns = FALSE,
cluster_row_slices = FALSE,
cluster_column_slices = FALSE,
show_row_names = FALSE,
show_column_names = FALSE,
row_names_side = ifelse(flip, "left", "right"),
column_names_side = ifelse(flip, "bottom", "top"),
row_names_rot = 0,
column_names_rot = 90,
row_title = NULL,
column_title = NULL,
row_title_side = "left",
column_title_side = "top",
row_title_rot = 0,
column_title_rot = ifelse(flip, 90, 0),
anno_terms = FALSE,
anno_keys = FALSE,
anno_features = FALSE,
terms_width = grid::unit(4, "in"),
terms_fontsize = 8,
keys_width = grid::unit(2, "in"),
keys_fontsize = c(6, 10),
features_width = grid::unit(2, "in"),
features_fontsize = c(6, 10),
IDtype = "symbol",
species = "Homo_sapiens",
db_update = FALSE,
db_version = "latest",
db_combine = FALSE,
convert_species = FALSE,
Ensembl_version = NULL,
mirror = NULL,
db = "GO_BP",
TERM2GENE = NULL,
TERM2NAME = NULL,
minGSSize = 10,
maxGSSize = 500,
GO_simplify = FALSE,
GO_simplify_cutoff = "p.adjust < 0.05",
simplify_method = "Wang",
simplify_similarityCutoff = 0.7,
pvalueCutoff = NULL,
padjustCutoff = 0.05,
topTerm = 5,
show_termid = FALSE,
topWord = 20,
words_excluded = NULL,
nlabel = 20,
features_label = NULL,
label_size = 10,
label_color = "black",
heatmap_palette = "RdBu",
heatmap_palcolor = NULL,
group_palette = "Paired",
group_palcolor = NULL,
cell_split_palette = "simspec",
cell_split_palcolor = NULL,
feature_split_palette = "simspec",
feature_split_palcolor = NULL,
cell_annotation = NULL,
cell_annotation_palette = "Paired",
cell_annotation_palcolor = NULL,
cell_annotation_params = if (flip) {
list(width = grid::unit(5, "mm"))
} else {
list(height = grid::unit(5, "mm"))
},
feature_annotation = NULL,
feature_annotation_palette = "Dark2",
feature_annotation_palcolor = NULL,
feature_annotation_params = if (flip) {
list(height = grid::unit(5, "mm"))
} else
{
list(width = grid::unit(5, "mm"))
},
use_raster = NULL,
raster_device = "png",
raster_by_magick = FALSE,
height = NULL,
width = NULL,
units = "inch",
seed = 11,
ht_params = list(),
verbose = TRUE
)Arguments
- srt
A Seurat object.
- features
A character vector of features to use. Default is
NULL.- cells
A character vector of cell names to use. Default is
NULL.- group.by
Name of one or more meta.data columns to group (color) cells by.
- split.by
Name of a column in meta.data column to split plot by. Default is
NULL.- within_groups
Whether to create separate heatmap scales for each group or within each group. Default is
FALSE.- max_cells
An integer, maximum number of cells to sample per group. Default is
100.- cell_order
A vector of cell names defining the order of cells. Default is
NULL.- border
Whether to add a border to the heatmap. Default is
TRUE.- flip
Whether to flip the heatmap. Default is
FALSE.- layer
Which layer to use. Default is
"counts".- assay
Which assay to use. If
NULL, the default assay of the Seurat object will be used.- exp_method
A character vector specifying the method for calculating expression values. Options are
"zscore","raw","fc","log2fc", or"log1p". Default is"zscore".- exp_legend_title
A character vector specifying the title for the legend of expression value. Default is
NULL.- limits
A two-length numeric vector specifying the limits for the color scale. Default is
NULL.- lib_normalize
Whether to normalize the data by library size.
- libsize
A numeric vector specifying the library size for each cell. Default is
NULL.- feature_split
A factor specifying how to split the features. Default is
NULL.- feature_split_by
A character vector specifying which group.by to use when splitting features (into n_split feature clusters). Default is
NULL.- n_split
A number of feature splits (feature clusters) to create. Default is
NULL.- split_order
A numeric vector specifying the order of splits. Default is
NULL.- split_method
A character vector specifying the method for splitting features. Options are
"kmeans","hclust", or"mfuzz". Default is"kmeans".- decreasing
Whether to sort feature splits in decreasing order. Default is
FALSE.- fuzzification
The fuzzification coefficient. Default is
NULL.- cluster_features_by
A character vector specifying which group.by to use when clustering features. Default is
NULL. By default, this parameter is set to NULL, which means that all groups will be used.- cluster_rows
Whether to cluster rows in the heatmap. Default is
FALSE.- cluster_columns
Whether to cluster columns in the heatmap. Default is
FALSE.- cluster_row_slices
Whether to cluster row slices in the heatmap. Default is
FALSE.- cluster_column_slices
Whether to cluster column slices in the heatmap. Default is
FALSE.- show_row_names
Whether to show row names in the heatmap. Default is
FALSE.- show_column_names
Whether to show column names in the heatmap. Default is
FALSE.- row_names_side
A character vector specifying the side to place row names.
- column_names_side
A character vector specifying the side to place column names.
- row_names_rot
The rotation angle for row names. Default is
0.- column_names_rot
The rotation angle for column names. Default is
90.- row_title
A character vector specifying the title for rows. Default is
NULL.- column_title
A character vector specifying the title for columns. Default is
NULL.- row_title_side
A character vector specifying the side to place row title. Default is
"left".- column_title_side
A character vector specifying the side to place column title. Default is
"top".- row_title_rot
The rotation angle for row title. Default is
0.- column_title_rot
The rotation angle for column title.
- anno_terms
Whether to include term annotations. Default is
FALSE.- anno_keys
Whether to include key annotations. Default is
FALSE.- anno_features
Whether to include feature annotations. Default is
FALSE.- terms_width
A unit specifying the width of term annotations. Default is
unit(4, "in").- terms_fontsize
A numeric vector specifying the font size(s) for term annotations. Default is
8.- keys_width
A unit specifying the width of key annotations. Default is
unit(2, "in").- keys_fontsize
A two-length numeric vector specifying the minimum and maximum font size(s) for key annotations. Default is
c(6, 10).- features_width
A unit specifying the width of feature annotations. Default is
unit(2, "in").- features_fontsize
A two-length numeric vector specifying the minimum and maximum font size(s) for feature annotations. Default is
c(6, 10).- IDtype
A character vector specifying the type of IDs for features. Default is
"symbol".- species
A character vector specifying the species for features. Default is
"Homo_sapiens".- db_update
Whether the gene annotation databases should be forcefully updated. If set to FALSE, the function will attempt to load the cached databases instead. Default is
FALSE.- db_version
A character vector specifying the version of the gene annotation databases to be retrieved. Default is
"latest".- db_combine
Whether to use a combined database. Default is
FALSE.- convert_species
Whether to use a species-converted database when the annotation is missing for the specified species. Default is
TRUE.- Ensembl_version
An integer specifying the Ensembl version. Default is
NULL. IfNULL, the latest version will be used.- mirror
A character vector specifying the mirror for the Ensembl database. Default is
NULL.- db
A character vector specifying the database to use. Default is
"GO_BP".- TERM2GENE
A data.frame specifying the TERM2GENE mapping for the database. Default is
NULL.- TERM2NAME
A data.frame specifying the TERM2NAME mapping for the database. Default is
NULL.- minGSSize
An integer specifying the minimum gene set size for the database. Default is
10.- maxGSSize
An integer specifying the maximum gene set size for the database. Default is
500.- GO_simplify
Whether to simplify gene ontology terms. Default is
FALSE.- GO_simplify_cutoff
A character vector specifying the cutoff for GO simplification. Default is
"p.adjust < 0.05".- simplify_method
A character vector specifying the method for GO simplification. Default is
"Wang".- simplify_similarityCutoff
The similarity cutoff for GO simplification. Default is
0.7.- pvalueCutoff
A numeric vector specifying the p-value cutoff(s) for significance. Default is
NULL.- padjustCutoff
The adjusted p-value cutoff for significance. Default is
0.05.- topTerm
A number of top terms to include. Default is
5.- show_termid
Whether to show term IDs. Default is
FALSE.- topWord
A number of top words to include. Default is
20.- words_excluded
A character vector specifying the words to exclude. Default is
NULL.- nlabel
A number of labels to include. Default is
20.- features_label
A character vector specifying the features to label. Default is
NULL.- label_size
The size of labels. Default is
10.- label_color
A character vector specifying the color of labels. Default is
"black".- heatmap_palette
A character vector specifying the palette to use for the heatmap. Default is
"RdBu".- heatmap_palcolor
A character vector specifying the heatmap color to use. Default is
NULL.- group_palette
A character vector specifying the palette to use for groups. Default is
"Paired".- group_palcolor
A character vector specifying the group color to use. Default is
NULL.- cell_split_palette
A character vector specifying the palette to use for cell splits. Default is
"simspec".- cell_split_palcolor
A character vector specifying the cell split color to use. Default is
NULL.- feature_split_palette
A character vector specifying the palette to use for feature splits. Default is
"simspec".- feature_split_palcolor
A character vector specifying the feature split color to use. Default is
NULL.- cell_annotation
A character vector specifying the cell annotation(s) to include. Default is
NULL.- cell_annotation_palette
A character vector specifying the palette to use for cell annotations. The length of the vector should match the number of cell_annotation. Default is
"Paired".- cell_annotation_palcolor
A list of character vector specifying the cell annotation color(s) to use. The length of the list should match the number of cell_annotation. Default is
NULL.- cell_annotation_params
A list specifying additional parameters for cell annotations. Default is a list with
width = unit(1, "cm")if flip is TRUE, else a list withheight = unit(1, "cm").- feature_annotation
A character vector specifying the feature annotation(s) to include. Default is
NULL.- feature_annotation_palette
A character vector specifying the palette to use for feature annotations. The length of the vector should match the number of feature_annotation. Default is
"Dark2".- feature_annotation_palcolor
A list of character vector specifying the feature annotation color to use. The length of the list should match the number of feature_annotation. Default is
NULL.- feature_annotation_params
A list specifying additional parameters for feature annotations. Default is
list().- use_raster
Whether to use a raster device for plotting. Default is
NULL.- raster_device
A character vector specifying the raster device to use. Default is
"png".- raster_by_magick
Whether to use the 'magick' package for raster. Default is
FALSE.- height
A numeric vector specifying the height(s) of the heatmap body. Default is
NULL.- width
A numeric vector specifying the width(s) of the heatmap body. Default is
NULL.- units
A character vector specifying the units for the height and width. Default is
"inch".- seed
Random seed for reproducibility. Default is
11.- ht_params
Additional parameters to customize the appearance of the heatmap. This should be a list with named elements, where the names correspond to parameter names in the ComplexHeatmap::Heatmap function. Any conflicting parameters will override the defaults set by this function. Default is
list().- verbose
Whether to print the message. Default is
TRUE.
Examples
data(pancreas_sub)
pancreas_sub <- standard_scop(pancreas_sub)
#> ℹ [2026-02-11 03:32:31] Start standard scop workflow...
#> ℹ [2026-02-11 03:32:32] Checking a list of <Seurat>...
#> ! [2026-02-11 03:32:32] Data 1/1 of the `srt_list` is "unknown"
#> ℹ [2026-02-11 03:32:32] Perform `NormalizeData()` with `normalization.method = 'LogNormalize'` on the data 1/1 of the `srt_list`...
#> ℹ [2026-02-11 03:32:34] Perform `Seurat::FindVariableFeatures()` on the data 1/1 of the `srt_list`...
#> ℹ [2026-02-11 03:32:34] Use the separate HVF from srt_list
#> ℹ [2026-02-11 03:32:34] Number of available HVF: 2000
#> ℹ [2026-02-11 03:32:34] Finished check
#> ℹ [2026-02-11 03:32:35] Perform `Seurat::ScaleData()`
#> ℹ [2026-02-11 03:32:35] Perform pca linear dimension reduction
#> ℹ [2026-02-11 03:32:36] Perform `Seurat::FindClusters()` with `cluster_algorithm = 'louvain'` and `cluster_resolution = 0.6`
#> ℹ [2026-02-11 03:32:36] Reorder clusters...
#> ℹ [2026-02-11 03:32:36] Perform umap nonlinear dimension reduction
#> ℹ [2026-02-11 03:32:36] Non-linear dimensionality reduction (umap) using (Standardpca) dims (1-50) as input
#> ℹ [2026-02-11 03:32:39] Non-linear dimensionality reduction (umap) using (Standardpca) dims (1-50) as input
#> ✔ [2026-02-11 03:32:43] Run scop standard workflow completed
pancreas_sub <- RunDEtest(
pancreas_sub,
group.by = "CellType"
)
#> ℹ [2026-02-11 03:32:43] Data type is log-normalized
#> ℹ [2026-02-11 03:32:43] Start differential expression test
#> ℹ [2026-02-11 03:32:43] Find all markers(wilcox) among [1] 5 groups...
#> ℹ [2026-02-11 03:32:43] Using 1 core
#> ⠙ [2026-02-11 03:32:43] Running for Ductal [1/5] ■■■■■■■ …
#> ✔ [2026-02-11 03:32:43] Completed 5 tasks in 741ms
#>
#> ℹ [2026-02-11 03:32:43] Building results
#> ✔ [2026-02-11 03:32:44] Differential expression test completed
de_filter <- dplyr::filter(
pancreas_sub@tools$DEtest_CellType$AllMarkers_wilcox,
p_val_adj < 0.05 & avg_log2FC > 1
)
ht1 <- FeatureHeatmap(
pancreas_sub,
features = de_filter$gene,
group.by = "CellType",
split.by = "Phase",
cell_split_palette = "Dark2"
)
#> `use_raster` is automatically set to TRUE for a matrix with more than
#> 2000 rows. You can control `use_raster` argument by explicitly setting
#> TRUE/FALSE to it.
#>
#> Set `ht_opt$message = FALSE` to turn off this message.
ht1$plot
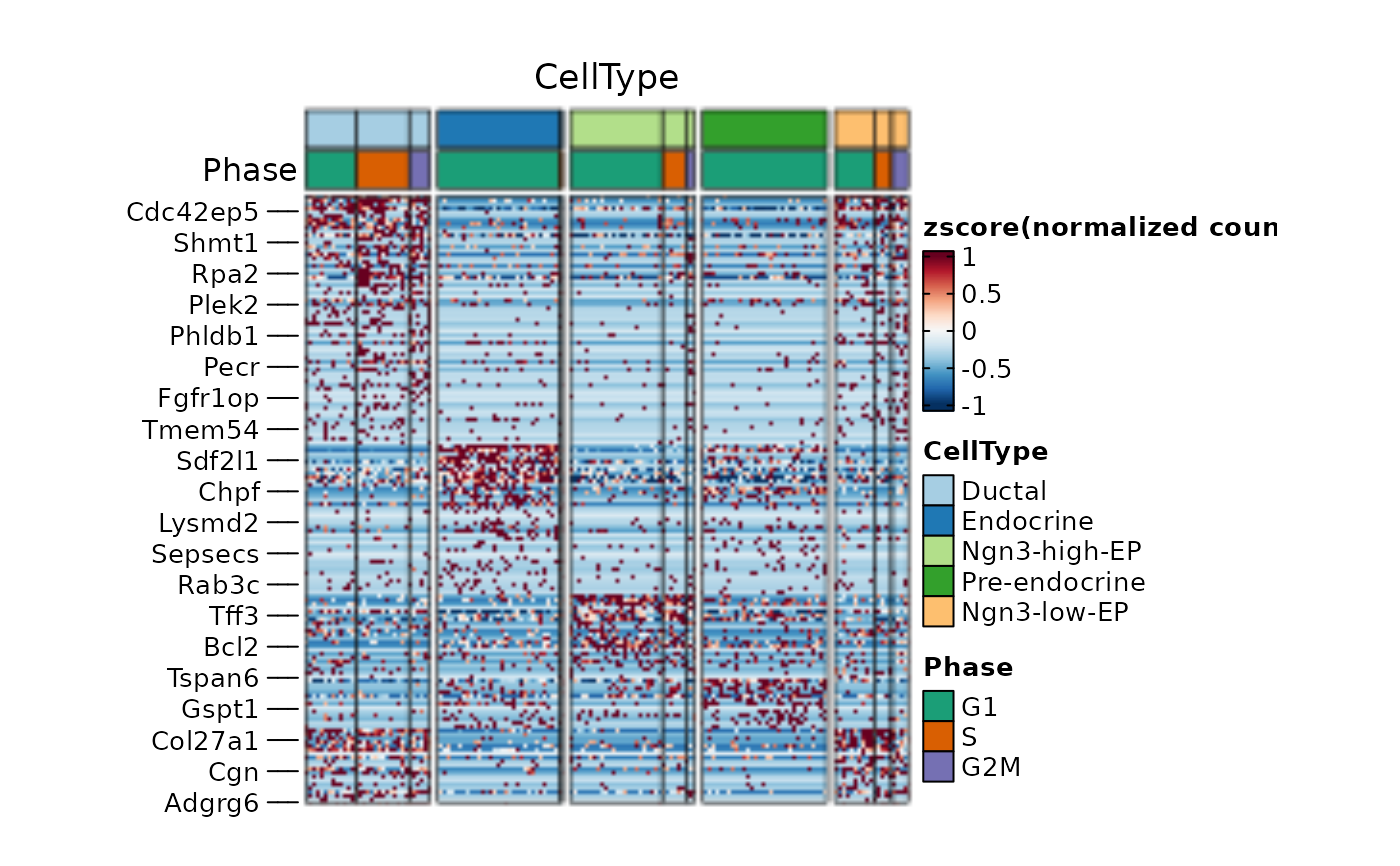
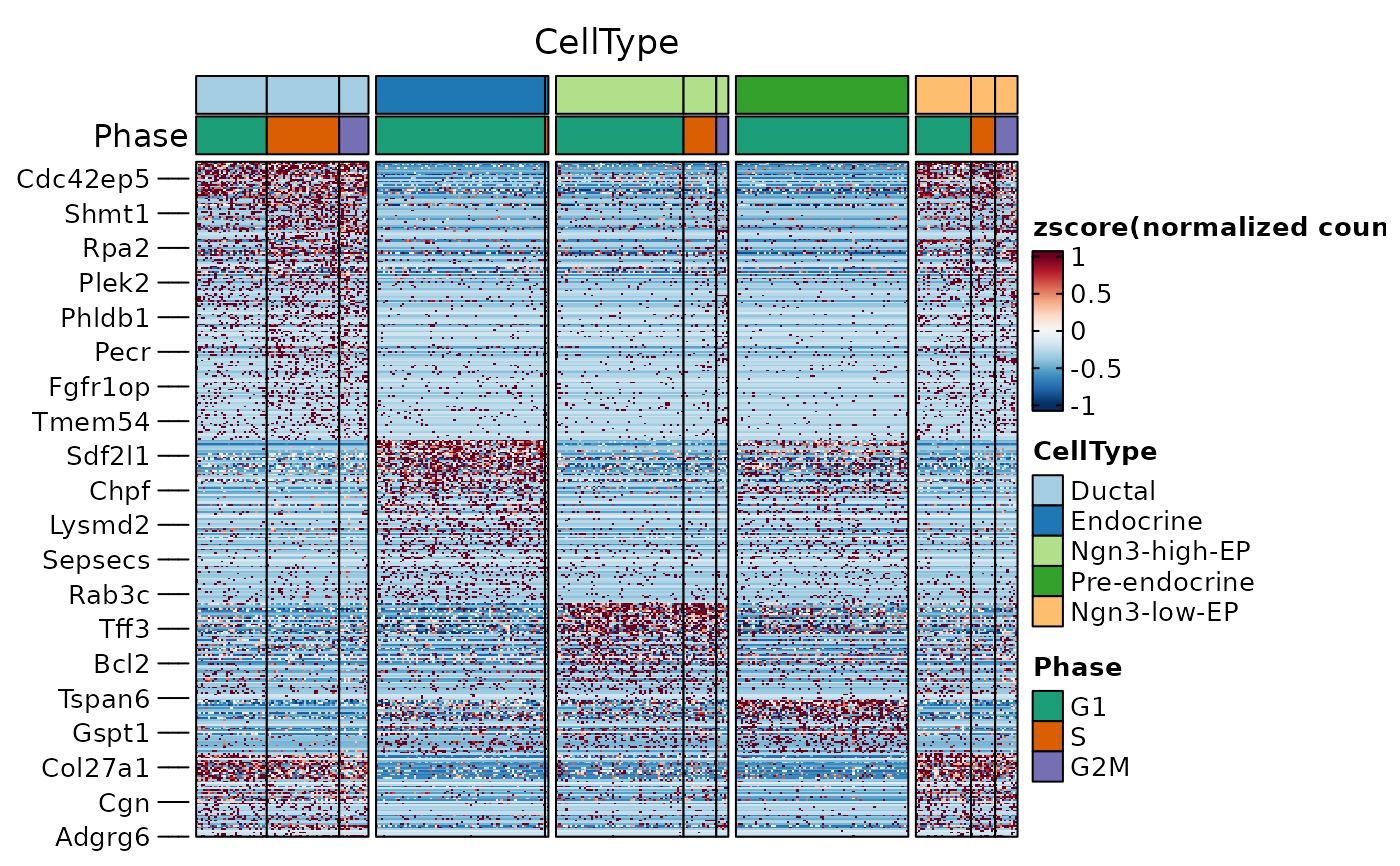 thisplot::panel_fix(
ht1$plot,
height = 4,
width = 6,
raster = TRUE,
dpi = 50
)
thisplot::panel_fix(
ht1$plot,
height = 4,
width = 6,
raster = TRUE,
dpi = 50
)
 ht2 <- FeatureHeatmap(
pancreas_sub,
features = de_filter$gene,
group.by = c("CellType", "SubCellType"),
n_split = 4,
cluster_rows = TRUE,
cluster_row_slices = TRUE,
cluster_columns = TRUE,
cluster_column_slices = TRUE,
ht_params = list(row_gap = grid::unit(0, "mm")),
use_raster = FALSE
)
#> ℹ [2026-02-11 03:32:57] The size of the heatmap is fixed because certain elements are not scalable.
#> ℹ [2026-02-11 03:32:57] The width and height of the heatmap are determined by the size of the current viewport.
#> ℹ [2026-02-11 03:32:57] If you want to have more control over the size, you can manually set the parameters 'width' and 'height'.
ht2 <- FeatureHeatmap(
pancreas_sub,
features = de_filter$gene,
group.by = c("CellType", "SubCellType"),
n_split = 4,
cluster_rows = TRUE,
cluster_row_slices = TRUE,
cluster_columns = TRUE,
cluster_column_slices = TRUE,
ht_params = list(row_gap = grid::unit(0, "mm")),
use_raster = FALSE
)
#> ℹ [2026-02-11 03:32:57] The size of the heatmap is fixed because certain elements are not scalable.
#> ℹ [2026-02-11 03:32:57] The width and height of the heatmap are determined by the size of the current viewport.
#> ℹ [2026-02-11 03:32:57] If you want to have more control over the size, you can manually set the parameters 'width' and 'height'.
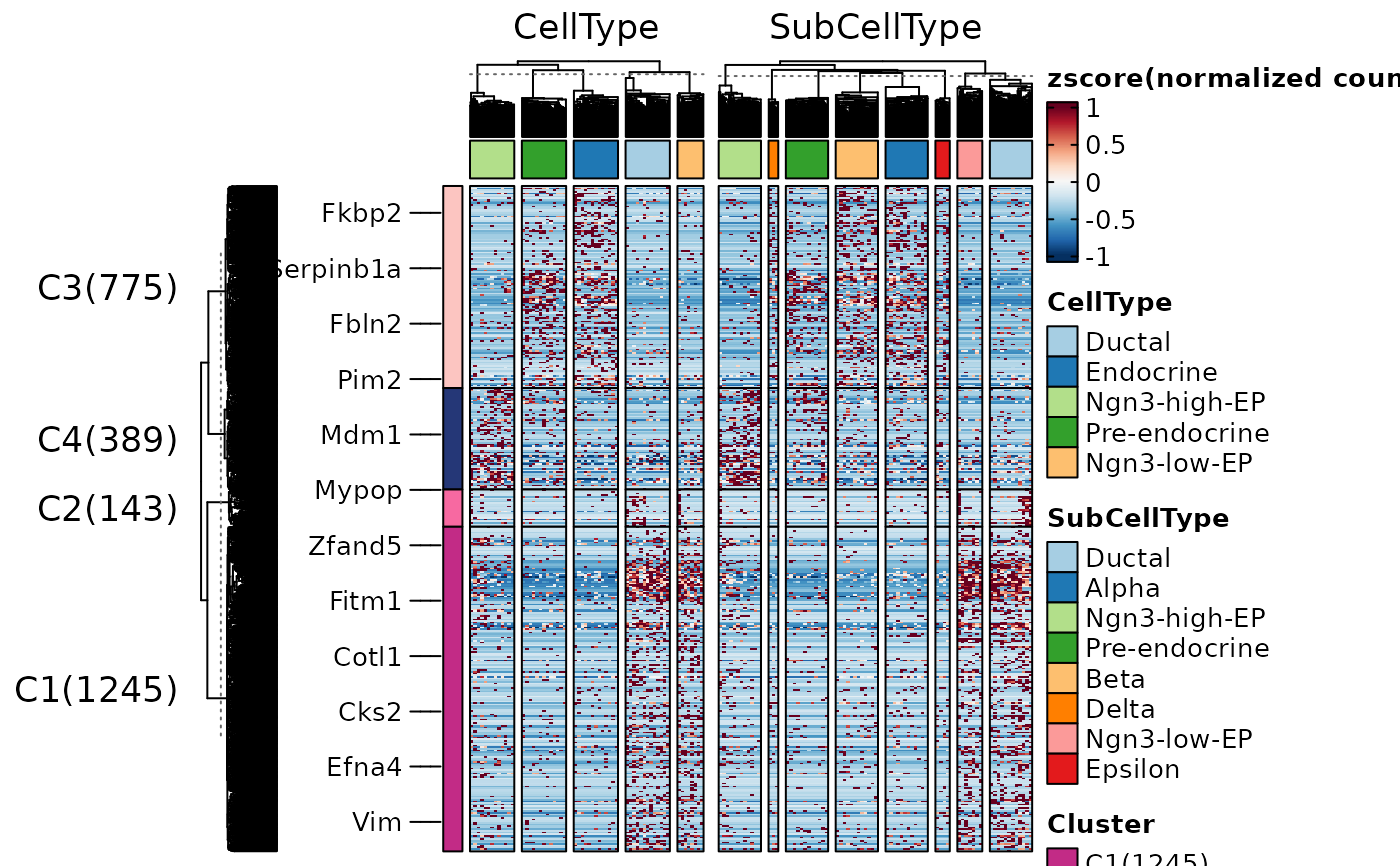 ht2$plot
ht2$plot
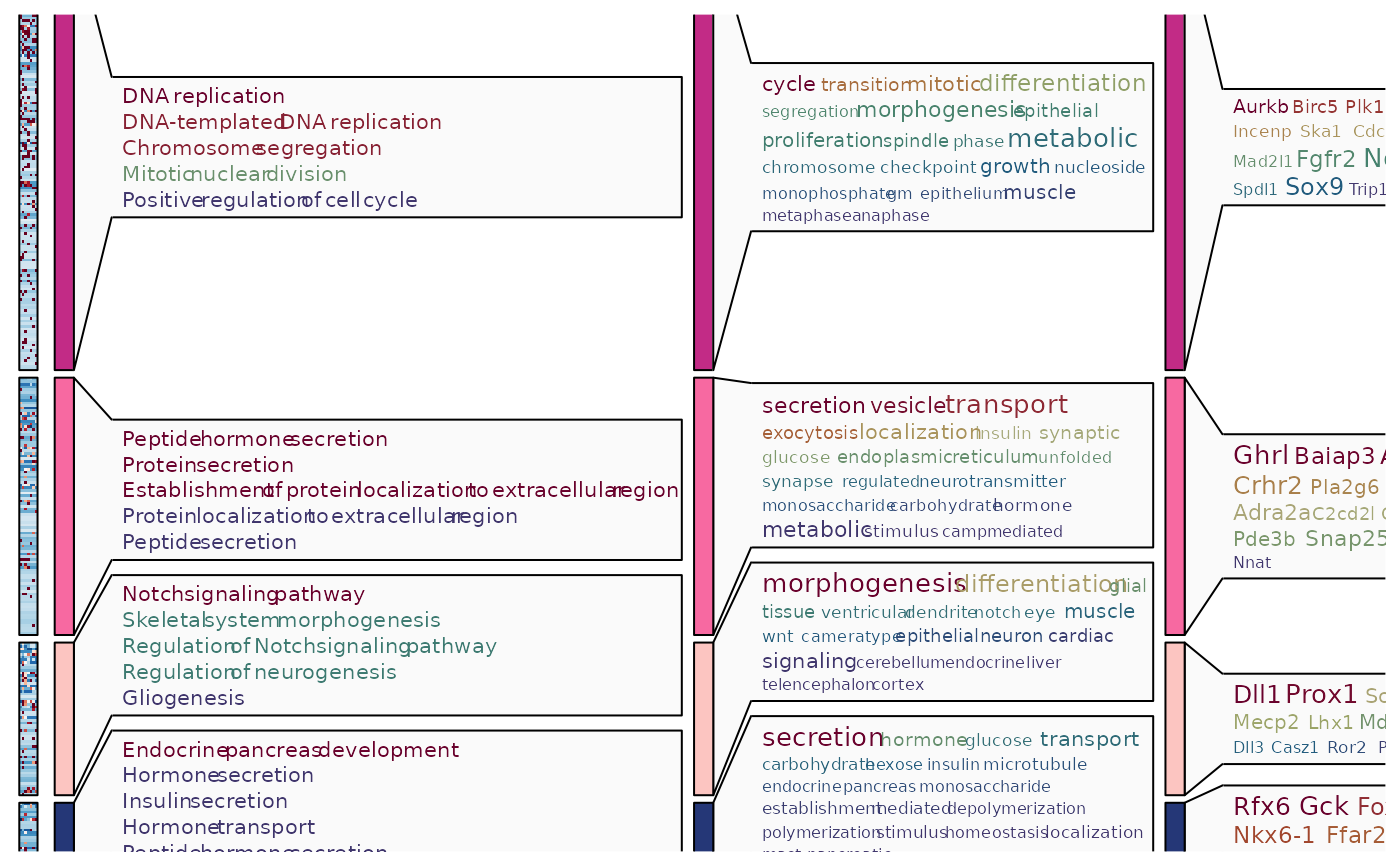
 ht3 <- FeatureHeatmap(
pancreas_sub,
features = de_filter$gene,
feature_split = de_filter$group1,
group.by = "CellType",
species = "Mus_musculus",
db = "GO_BP",
anno_terms = TRUE,
anno_keys = TRUE,
anno_features = TRUE
)
#> ℹ [2026-02-11 03:33:26] Start Enrichment analysis
#> ℹ [2026-02-11 03:33:26] Species: "Mus_musculus"
#> ℹ [2026-02-11 03:33:26] Loading cached: GO_BP version: 3.22.0 nterm:15169 created: 2026-02-11 03:27:59
#> ℹ [2026-02-11 03:33:27] Permform enrichment...
#> ℹ [2026-02-11 03:33:27] Using 1 core
#> ⠙ [2026-02-11 03:33:27] Running for 1 [1/5] ■■■■■■■ 2…
#> ⠹ [2026-02-11 03:33:27] Running for 2 [2/5] ■■■■■■■■■■■■■ 4…
#> ⠸ [2026-02-11 03:33:27] Running for 3 [3/5] ■■■■■■■■■■■■■■■■■■■ 6…
#> ⠼ [2026-02-11 03:33:27] Running for 4 [4/5] ■■■■■■■■■■■■■■■■■■■■■■■■■ 8…
#> ✔ [2026-02-11 03:33:27] Completed 5 tasks in 1m 16.1s
#>
#> ℹ [2026-02-11 03:33:27] Building results
#> ✔ [2026-02-11 03:34:44] Enrichment analysis done
#> `use_raster` is automatically set to TRUE for a matrix with more than
#> 2000 rows. You can control `use_raster` argument by explicitly setting
#> TRUE/FALSE to it.
#>
#> Set `ht_opt$message = FALSE` to turn off this message.
#> ℹ [2026-02-11 03:35:24] The size of the heatmap is fixed because certain elements are not scalable.
#> ℹ [2026-02-11 03:35:24] The width and height of the heatmap are determined by the size of the current viewport.
#> ℹ [2026-02-11 03:35:24] If you want to have more control over the size, you can manually set the parameters 'width' and 'height'.
ht3 <- FeatureHeatmap(
pancreas_sub,
features = de_filter$gene,
feature_split = de_filter$group1,
group.by = "CellType",
species = "Mus_musculus",
db = "GO_BP",
anno_terms = TRUE,
anno_keys = TRUE,
anno_features = TRUE
)
#> ℹ [2026-02-11 03:33:26] Start Enrichment analysis
#> ℹ [2026-02-11 03:33:26] Species: "Mus_musculus"
#> ℹ [2026-02-11 03:33:26] Loading cached: GO_BP version: 3.22.0 nterm:15169 created: 2026-02-11 03:27:59
#> ℹ [2026-02-11 03:33:27] Permform enrichment...
#> ℹ [2026-02-11 03:33:27] Using 1 core
#> ⠙ [2026-02-11 03:33:27] Running for 1 [1/5] ■■■■■■■ 2…
#> ⠹ [2026-02-11 03:33:27] Running for 2 [2/5] ■■■■■■■■■■■■■ 4…
#> ⠸ [2026-02-11 03:33:27] Running for 3 [3/5] ■■■■■■■■■■■■■■■■■■■ 6…
#> ⠼ [2026-02-11 03:33:27] Running for 4 [4/5] ■■■■■■■■■■■■■■■■■■■■■■■■■ 8…
#> ✔ [2026-02-11 03:33:27] Completed 5 tasks in 1m 16.1s
#>
#> ℹ [2026-02-11 03:33:27] Building results
#> ✔ [2026-02-11 03:34:44] Enrichment analysis done
#> `use_raster` is automatically set to TRUE for a matrix with more than
#> 2000 rows. You can control `use_raster` argument by explicitly setting
#> TRUE/FALSE to it.
#>
#> Set `ht_opt$message = FALSE` to turn off this message.
#> ℹ [2026-02-11 03:35:24] The size of the heatmap is fixed because certain elements are not scalable.
#> ℹ [2026-02-11 03:35:24] The width and height of the heatmap are determined by the size of the current viewport.
#> ℹ [2026-02-11 03:35:24] If you want to have more control over the size, you can manually set the parameters 'width' and 'height'.
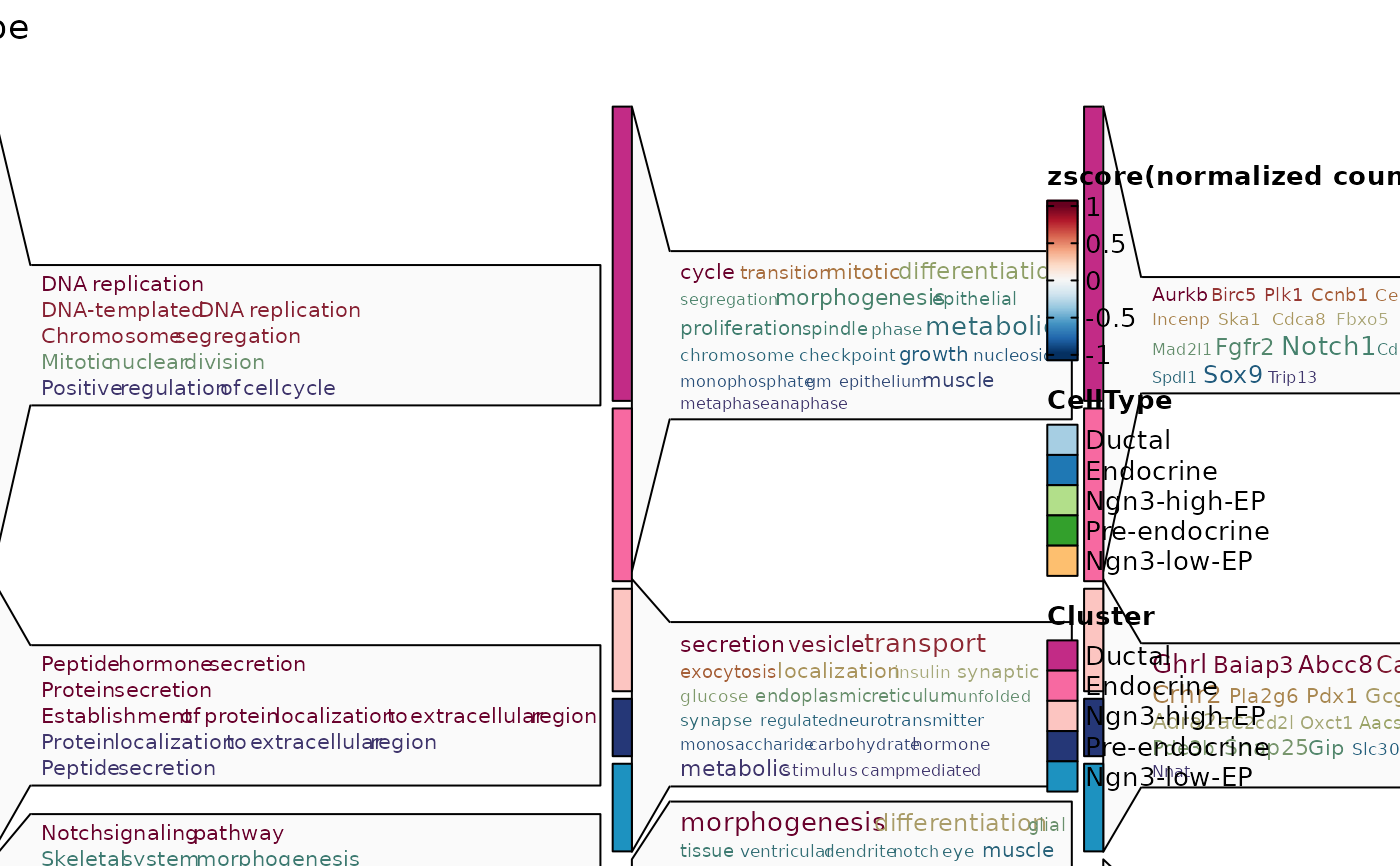 ht3$plot
ht3$plot
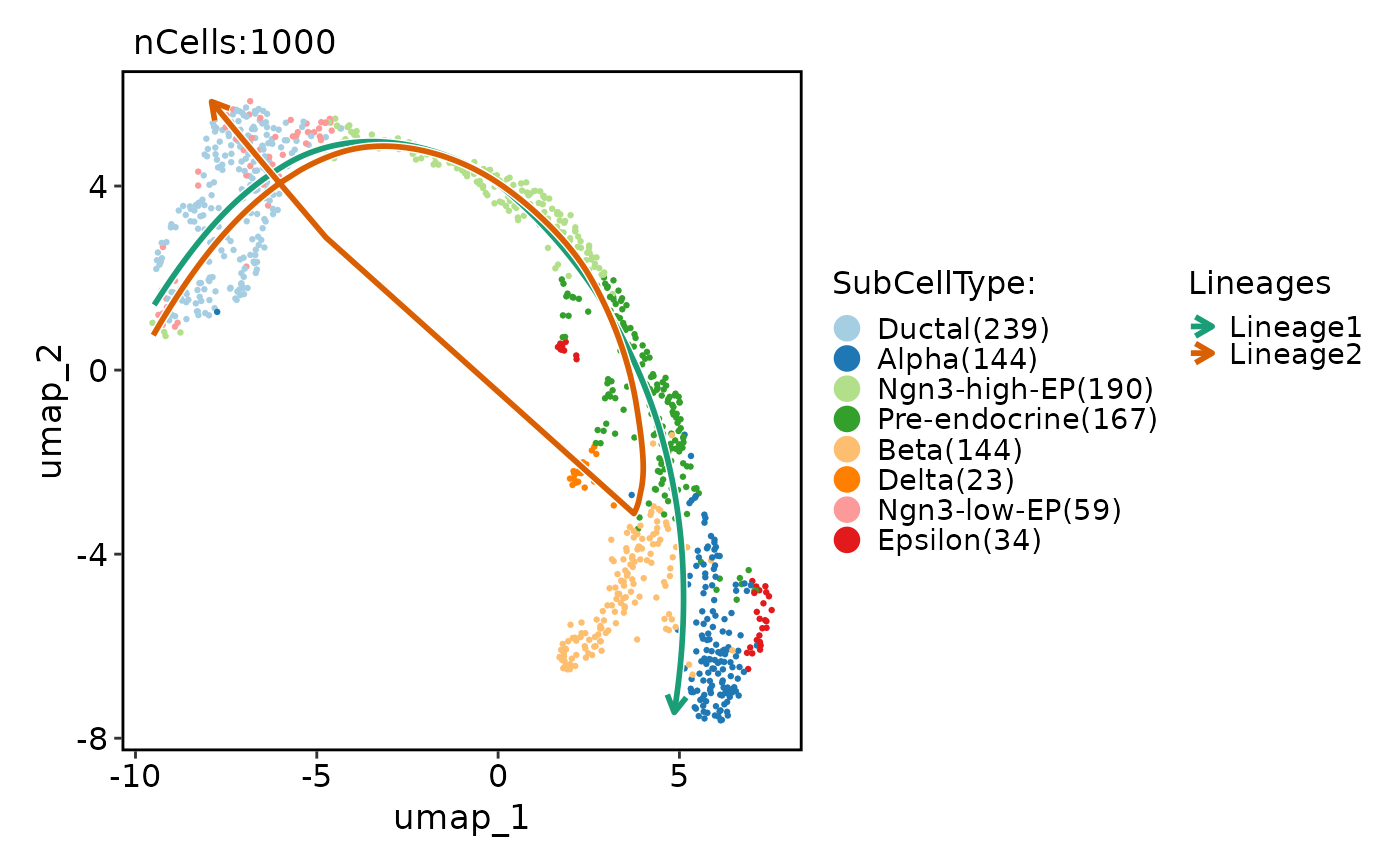
 pancreas_sub <- RunSlingshot(
pancreas_sub,
group.by = "SubCellType",
reduction = "UMAP"
)
#> Warning: No shared levels found between `names(values)` of the manual scale and the
#> data's fill values.
#> Warning: No shared levels found between `names(values)` of the manual scale and the
#> data's fill values.
#> Warning: Removed 3 rows containing missing values or values outside the scale range
#> (`geom_path()`).
#> Warning: Removed 3 rows containing missing values or values outside the scale range
#> (`geom_path()`).
pancreas_sub <- RunSlingshot(
pancreas_sub,
group.by = "SubCellType",
reduction = "UMAP"
)
#> Warning: No shared levels found between `names(values)` of the manual scale and the
#> data's fill values.
#> Warning: No shared levels found between `names(values)` of the manual scale and the
#> data's fill values.
#> Warning: Removed 3 rows containing missing values or values outside the scale range
#> (`geom_path()`).
#> Warning: Removed 3 rows containing missing values or values outside the scale range
#> (`geom_path()`).
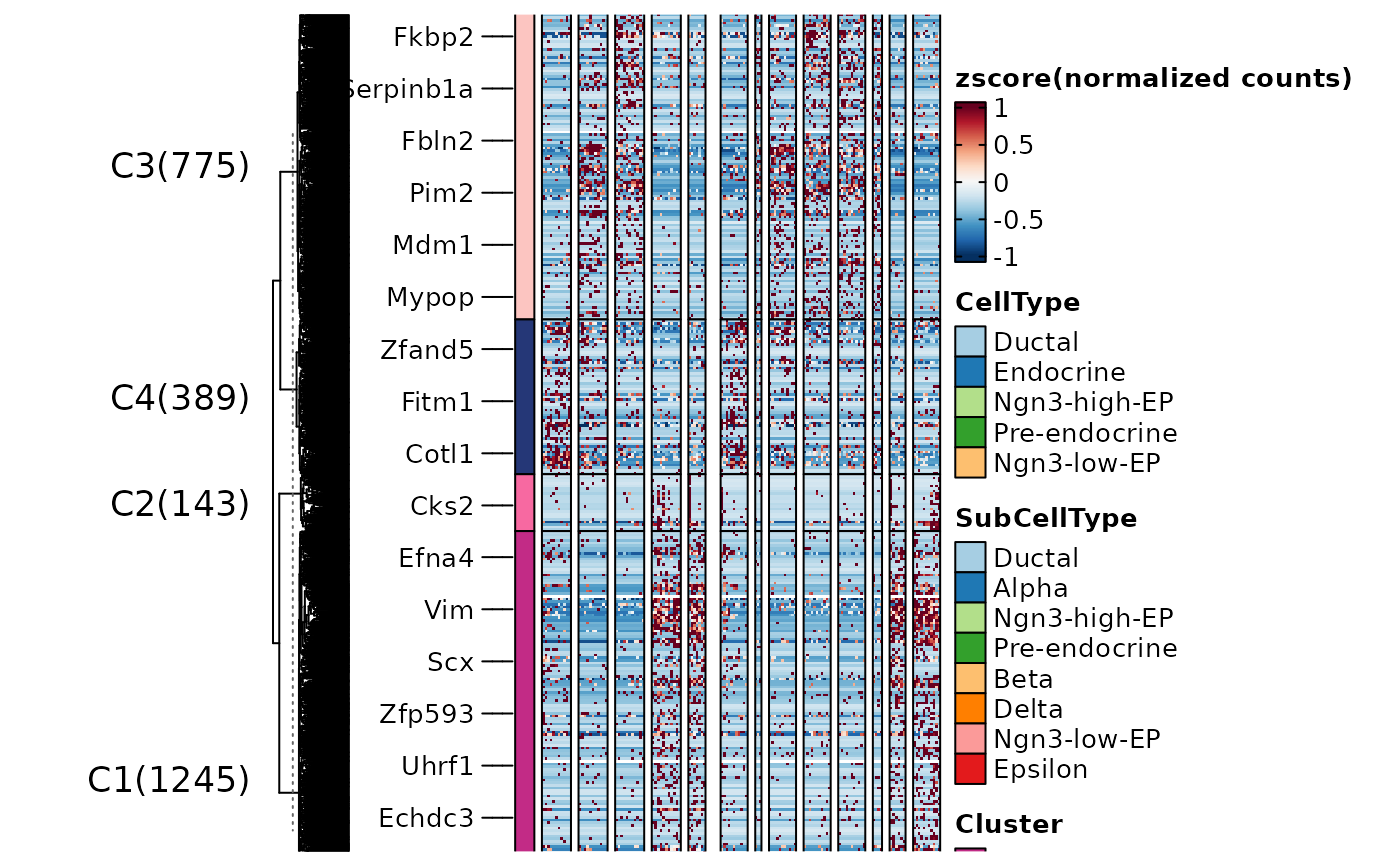
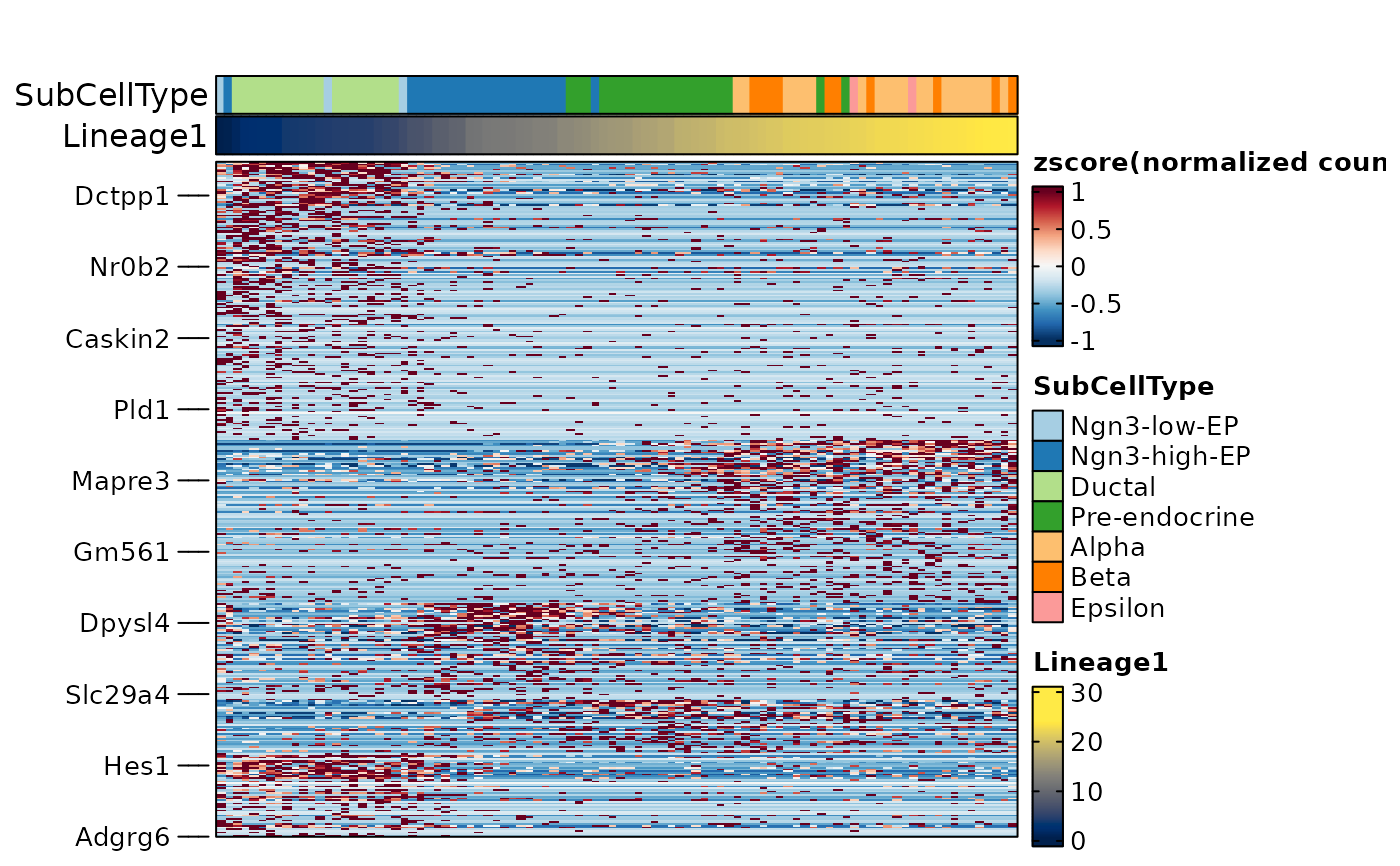
 ht4 <- FeatureHeatmap(
pancreas_sub,
features = de_filter$gene,
nlabel = 10,
cell_order = names(sort(pancreas_sub$Lineage1)),
cell_annotation = c("SubCellType", "Lineage1"),
cell_annotation_palette = c("Paired", "cividis")
)
#> `use_raster` is automatically set to TRUE for a matrix with more than
#> 2000 rows. You can control `use_raster` argument by explicitly setting
#> TRUE/FALSE to it.
#>
#> Set `ht_opt$message = FALSE` to turn off this message.
ht4$plot
ht4 <- FeatureHeatmap(
pancreas_sub,
features = de_filter$gene,
nlabel = 10,
cell_order = names(sort(pancreas_sub$Lineage1)),
cell_annotation = c("SubCellType", "Lineage1"),
cell_annotation_palette = c("Paired", "cividis")
)
#> `use_raster` is automatically set to TRUE for a matrix with more than
#> 2000 rows. You can control `use_raster` argument by explicitly setting
#> TRUE/FALSE to it.
#>
#> Set `ht_opt$message = FALSE` to turn off this message.
ht4$plot
 if (FALSE) { # \dontrun{
pancreas_sub <- AnnotateFeatures(
pancreas_sub,
species = "Mus_musculus",
db = c("CSPA", "TF")
)
ht5 <- FeatureHeatmap(
pancreas_sub,
features = de_filter$gene,
n_split = 4,
group.by = "CellType",
heatmap_palette = "viridis",
feature_annotation = c("TF", "CSPA"),
feature_annotation_palcolor = list(
c("gold", "steelblue"), c("forestgreen")
),
cell_annotation = c("Phase", "G2M_score"),
cell_annotation_palette = c("Dark2", "Purples")
)
ht5$plot
ht6 <- FeatureHeatmap(
pancreas_sub,
features = de_filter$gene,
n_split = 4,
group.by = "CellType",
heatmap_palette = "viridis",
feature_annotation = c("TF", "CSPA"),
feature_annotation_palcolor = list(
c("gold", "steelblue"), c("forestgreen")
),
cell_annotation = c("Phase", "G2M_score"),
cell_annotation_palette = c("Dark2", "Purples"),
flip = TRUE,
column_title_rot = 45
)
ht6$plot
} # }
if (FALSE) { # \dontrun{
pancreas_sub <- AnnotateFeatures(
pancreas_sub,
species = "Mus_musculus",
db = c("CSPA", "TF")
)
ht5 <- FeatureHeatmap(
pancreas_sub,
features = de_filter$gene,
n_split = 4,
group.by = "CellType",
heatmap_palette = "viridis",
feature_annotation = c("TF", "CSPA"),
feature_annotation_palcolor = list(
c("gold", "steelblue"), c("forestgreen")
),
cell_annotation = c("Phase", "G2M_score"),
cell_annotation_palette = c("Dark2", "Purples")
)
ht5$plot
ht6 <- FeatureHeatmap(
pancreas_sub,
features = de_filter$gene,
n_split = 4,
group.by = "CellType",
heatmap_palette = "viridis",
feature_annotation = c("TF", "CSPA"),
feature_annotation_palcolor = list(
c("gold", "steelblue"), c("forestgreen")
),
cell_annotation = c("Phase", "G2M_score"),
cell_annotation_palette = c("Dark2", "Purples"),
flip = TRUE,
column_title_rot = 45
)
ht6$plot
} # }