Run cell-level quality control for single cell RNA-seq data.
Usage
RunCellQC(
srt,
assay = "RNA",
split.by = NULL,
return_filtered = FALSE,
qc_metrics = c("doublets", "outlier", "umi", "gene", "mito", "ribo", "ribo_mito_ratio",
"species"),
db_method = "scDblFinder",
db_rate = NULL,
db_coefficient = 0.01,
outlier_threshold = c("log10_nCount:lower:2.5", "log10_nCount:higher:5",
"log10_nFeature:lower:2.5", "log10_nFeature:higher:5", "featurecount_dist:lower:2.5"),
outlier_n = 1,
UMI_threshold = 3000,
gene_threshold = 1000,
mito_threshold = 20,
mito_pattern = c("MT-", "Mt-", "mt-"),
mito_gene = NULL,
ribo_threshold = 50,
ribo_pattern = c("RP[SL]\\d+\\w{0,1}\\d*$", "Rp[sl]\\d+\\w{0,1}\\d*$",
"rp[sl]\\d+\\w{0,1}\\d*$"),
ribo_gene = NULL,
ribo_mito_ratio_range = c(1, Inf),
species = NULL,
species_gene_prefix = NULL,
species_percent = 95,
seed = 11
)Arguments
- srt
A Seurat object.
- assay
The name of the assay to be used for doublet-calling. Default is
"RNA".- split.by
Name of a column in meta.data column to split plot by. Default is
NULL.- return_filtered
Logical indicating whether to return a cell-filtered Seurat object. Default is
FALSE.- qc_metrics
A character vector specifying the quality control metrics to be applied. Default is
c("doublets", "outlier", "umi", "gene", "mito", "ribo", "ribo_mito_ratio", "species").- db_method
Method used for doublet-calling. Can be one of
"scDblFinder","Scrublet","DoubletDetection","scds_cxds","scds_bcds","scds_hybrid".- db_rate
The expected doublet rate. Default is calculated as
ncol(srt) / 1000 * 0.01.- db_coefficient
The coefficient used to calculate the doublet rate. Default is
0.01. Doublet rate is calculated asncol(srt) / 1000 * db_coefficient.- outlier_threshold
A character vector specifying the outlier threshold. Default is
c("log10_nCount:lower:2.5", "log10_nCount:higher:5", "log10_nFeature:lower:2.5", "log10_nFeature:higher:5", "featurecount_dist:lower:2.5"). See scuttle::isOutlier.- outlier_n
Minimum number of outlier metrics that meet the conditions for determining outlier cells. Default is
1.- UMI_threshold
UMI number threshold. Cells that exceed this threshold will be considered as kept. Default is
3000.- gene_threshold
Gene number threshold. Cells that exceed this threshold will be considered as kept. Default is
1000.- mito_threshold
Percentage of UMI counts of mitochondrial genes. Cells that exceed this threshold will be considered as discarded. Default is
20.- mito_pattern
Regex patterns to match the mitochondrial genes. Default is
c("MT-", "Mt-", "mt-").- mito_gene
A defined mitochondrial genes. If features provided, will ignore the
mito_patternmatching. Default isNULL.- ribo_threshold
Percentage of UMI counts of ribosomal genes. Cells that exceed this threshold will be considered as discarded. Default is
50.- ribo_pattern
Regex patterns to match the ribosomal genes. Default is
c("RP[SL]\\d+\\w{0,1}\\d*$", "Rp[sl]\\d+\\w{0,1}\\d*$", "rp[sl]\\d+\\w{0,1}\\d*$").- ribo_gene
A defined ribosomal genes. If features provided, will ignore the
ribo_patternmatching. Default isNULL.- ribo_mito_ratio_range
A numeric vector specifying the range of ribosomal/mitochondrial gene expression ratios for ribo_mito_ratio outlier cells. Default is
c(1, Inf).- species
Species used as the suffix of the QC metrics. The first is the species of interest. Default is
NULL.- species_gene_prefix
Species gene prefix used to calculate QC metrics for each species. Default is
NULL.- species_percent
Percentage of UMI counts of the first species. Cells that exceed this threshold will be considered as kept. Default is
95.- seed
Random seed for reproducibility. Default is
11.
Examples
data(pancreas_sub)
pancreas_sub <- standard_scop(pancreas_sub)
#> ℹ [2026-02-11 03:42:59] Start standard scop workflow...
#> ℹ [2026-02-11 03:43:00] Checking a list of <Seurat>...
#> ! [2026-02-11 03:43:00] Data 1/1 of the `srt_list` is "unknown"
#> ℹ [2026-02-11 03:43:00] Perform `NormalizeData()` with `normalization.method = 'LogNormalize'` on the data 1/1 of the `srt_list`...
#> ℹ [2026-02-11 03:43:02] Perform `Seurat::FindVariableFeatures()` on the data 1/1 of the `srt_list`...
#> ℹ [2026-02-11 03:43:02] Use the separate HVF from srt_list
#> ℹ [2026-02-11 03:43:02] Number of available HVF: 2000
#> ℹ [2026-02-11 03:43:03] Finished check
#> ℹ [2026-02-11 03:43:03] Perform `Seurat::ScaleData()`
#> ℹ [2026-02-11 03:43:03] Perform pca linear dimension reduction
#> ℹ [2026-02-11 03:43:04] Perform `Seurat::FindClusters()` with `cluster_algorithm = 'louvain'` and `cluster_resolution = 0.6`
#> ℹ [2026-02-11 03:43:04] Reorder clusters...
#> ℹ [2026-02-11 03:43:04] Perform umap nonlinear dimension reduction
#> ℹ [2026-02-11 03:43:04] Non-linear dimensionality reduction (umap) using (Standardpca) dims (1-50) as input
#> ℹ [2026-02-11 03:43:08] Non-linear dimensionality reduction (umap) using (Standardpca) dims (1-50) as input
#> ✔ [2026-02-11 03:43:12] Run scop standard workflow completed
pancreas_sub <- RunCellQC(pancreas_sub)
#> ℹ [2026-02-11 03:43:12] Data type is raw counts
#> ℹ [2026-02-11 03:43:12] Data type is raw counts
#> ℹ [2026-02-11 03:43:13] Data type is raw counts
#> ℹ [2026-02-11 03:47:58] >>> Total cells: [1] 1000
#> ℹ [2026-02-11 03:47:58] >>> Cells which are filtered out: [1] 45
#> ℹ [2026-02-11 03:47:58] >>> [1] 22 potential doublets
#> ℹ [2026-02-11 03:47:58] >>> [1] 23 outlier cells
#> ℹ [2026-02-11 03:47:58] >>> [1] 0low-UMI cells
#> ℹ [2026-02-11 03:47:58] >>> [1] 0low-gene cells
#> ℹ [2026-02-11 03:47:58] >>> [1] 0high-mito cells
#> ℹ [2026-02-11 03:47:58] >>> [1] 0high-ribo cells
#> ℹ [2026-02-11 03:47:58] >>> [1] 0ribo_mito_ratio outlier cells
#> ℹ [2026-02-11 03:47:58] >>> [1] 0species-contaminated cells
#> ℹ [2026-02-11 03:47:58] >>> Remained cells after filtering: [1] 955
CellStatPlot(
pancreas_sub,
stat.by = c(
"db_qc", "outlier_qc",
"umi_qc", "gene_qc",
"mito_qc", "ribo_qc",
"ribo_mito_ratio_qc", "species_qc"
),
plot_type = "upset",
stat_level = "Fail"
)
#> ! [2026-02-11 03:47:58] `stat_type` is forcibly set to "count" when plot "sankey", "chord", "venn", and "upset"
#> `geom_line()`: Each group consists of only one observation.
#> ℹ Do you need to adjust the group aesthetic?
#> `geom_line()`: Each group consists of only one observation.
#> ℹ Do you need to adjust the group aesthetic?
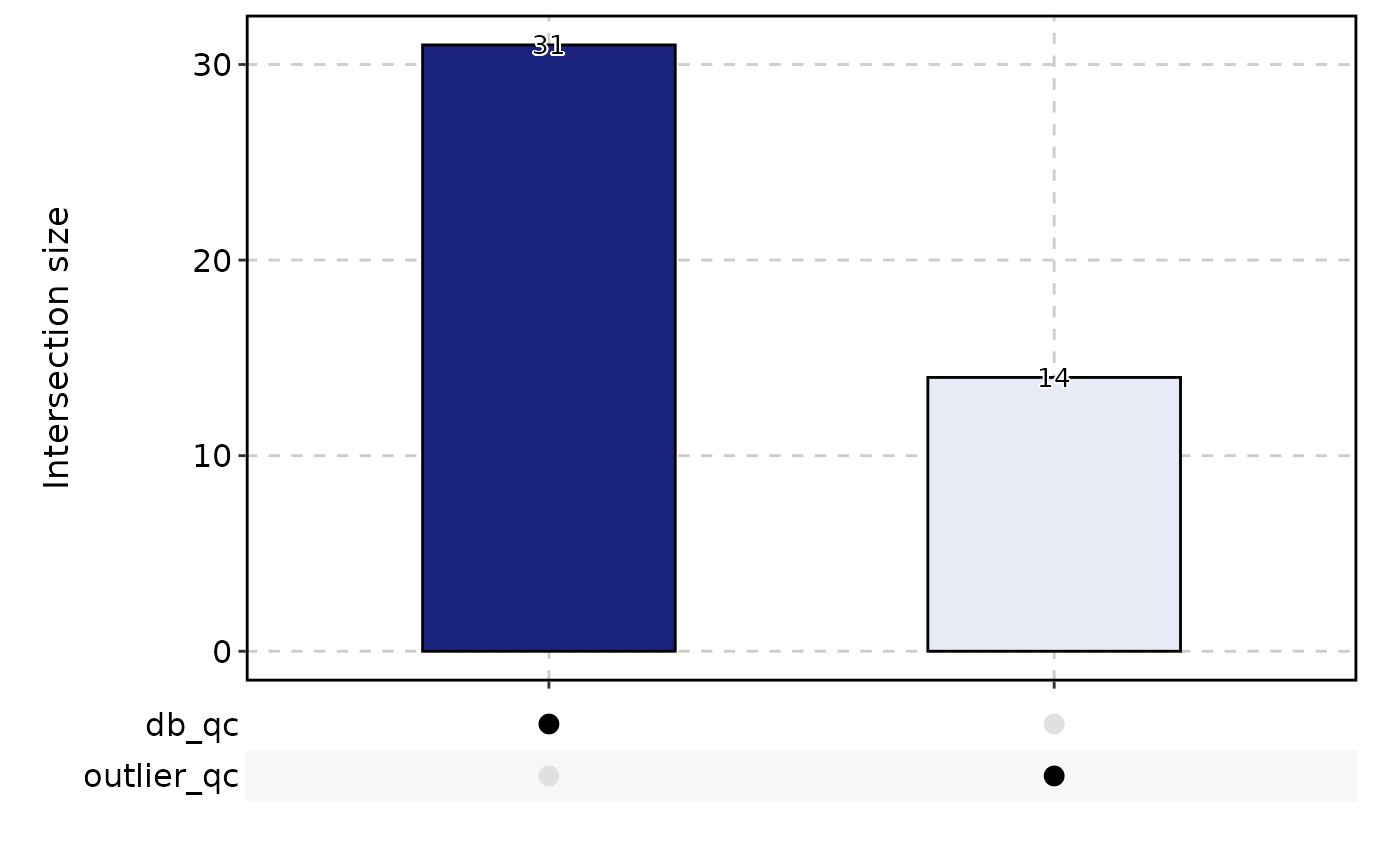 table(pancreas_sub$CellQC)
#>
#> Pass Fail
#> 955 45
data(ifnb_sub)
ifnb_sub <- RunCellQC(
srt = ifnb_sub,
split.by = "stim",
UMI_threshold = 1000,
gene_threshold = 550
)
#> ℹ [2026-02-11 03:47:59] Data type is raw counts
#> ℹ [2026-02-11 03:47:59] Running QC for CTRL
#> ℹ [2026-02-11 03:47:59] Data type is raw counts
#> ℹ [2026-02-11 03:47:59] Data type is raw counts
#> ℹ [2026-02-11 03:48:05] >>> Total cells: [1] 1000
#> ℹ [2026-02-11 03:48:05] >>> Cells which are filtered out: [1] 310
#> ℹ [2026-02-11 03:48:05] >>> [1] 49 potential doublets
#> ℹ [2026-02-11 03:48:05] >>> [1] 8 outlier cells
#> ℹ [2026-02-11 03:48:05] >>> [1] 28low-UMI cells
#> ℹ [2026-02-11 03:48:05] >>> [1] 250low-gene cells
#> ℹ [2026-02-11 03:48:05] >>> [1] 0high-mito cells
#> ℹ [2026-02-11 03:48:05] >>> [1] 0high-ribo cells
#> ℹ [2026-02-11 03:48:05] >>> [1] 0ribo_mito_ratio outlier cells
#> ℹ [2026-02-11 03:48:05] >>> [1] 0species-contaminated cells
#> ℹ [2026-02-11 03:48:05] >>> Remained cells after filtering: [1] 690
#> ℹ [2026-02-11 03:48:05] Running QC for STIM
#> ℹ [2026-02-11 03:48:05] Data type is raw counts
#> ℹ [2026-02-11 03:48:06] Data type is raw counts
#> ℹ [2026-02-11 03:48:12] >>> Total cells: [1] 1000
#> ℹ [2026-02-11 03:48:12] >>> Cells which are filtered out: [1] 308
#> ℹ [2026-02-11 03:48:12] >>> [1] 47 potential doublets
#> ℹ [2026-02-11 03:48:12] >>> [1] 12 outlier cells
#> ℹ [2026-02-11 03:48:12] >>> [1] 25low-UMI cells
#> ℹ [2026-02-11 03:48:12] >>> [1] 251low-gene cells
#> ℹ [2026-02-11 03:48:12] >>> [1] 0high-mito cells
#> ℹ [2026-02-11 03:48:12] >>> [1] 0high-ribo cells
#> ℹ [2026-02-11 03:48:12] >>> [1] 0ribo_mito_ratio outlier cells
#> ℹ [2026-02-11 03:48:12] >>> [1] 0species-contaminated cells
#> ℹ [2026-02-11 03:48:12] >>> Remained cells after filtering: [1] 692
CellStatPlot(
srt = ifnb_sub,
stat.by = c(
"db_qc", "outlier_qc",
"umi_qc", "gene_qc",
"mito_qc", "ribo_qc",
"ribo_mito_ratio_qc", "species_qc"
),
plot_type = "upset",
stat_level = "Fail"
)
#> ! [2026-02-11 03:48:12] `stat_type` is forcibly set to "count" when plot "sankey", "chord", "venn", and "upset"
table(pancreas_sub$CellQC)
#>
#> Pass Fail
#> 955 45
data(ifnb_sub)
ifnb_sub <- RunCellQC(
srt = ifnb_sub,
split.by = "stim",
UMI_threshold = 1000,
gene_threshold = 550
)
#> ℹ [2026-02-11 03:47:59] Data type is raw counts
#> ℹ [2026-02-11 03:47:59] Running QC for CTRL
#> ℹ [2026-02-11 03:47:59] Data type is raw counts
#> ℹ [2026-02-11 03:47:59] Data type is raw counts
#> ℹ [2026-02-11 03:48:05] >>> Total cells: [1] 1000
#> ℹ [2026-02-11 03:48:05] >>> Cells which are filtered out: [1] 310
#> ℹ [2026-02-11 03:48:05] >>> [1] 49 potential doublets
#> ℹ [2026-02-11 03:48:05] >>> [1] 8 outlier cells
#> ℹ [2026-02-11 03:48:05] >>> [1] 28low-UMI cells
#> ℹ [2026-02-11 03:48:05] >>> [1] 250low-gene cells
#> ℹ [2026-02-11 03:48:05] >>> [1] 0high-mito cells
#> ℹ [2026-02-11 03:48:05] >>> [1] 0high-ribo cells
#> ℹ [2026-02-11 03:48:05] >>> [1] 0ribo_mito_ratio outlier cells
#> ℹ [2026-02-11 03:48:05] >>> [1] 0species-contaminated cells
#> ℹ [2026-02-11 03:48:05] >>> Remained cells after filtering: [1] 690
#> ℹ [2026-02-11 03:48:05] Running QC for STIM
#> ℹ [2026-02-11 03:48:05] Data type is raw counts
#> ℹ [2026-02-11 03:48:06] Data type is raw counts
#> ℹ [2026-02-11 03:48:12] >>> Total cells: [1] 1000
#> ℹ [2026-02-11 03:48:12] >>> Cells which are filtered out: [1] 308
#> ℹ [2026-02-11 03:48:12] >>> [1] 47 potential doublets
#> ℹ [2026-02-11 03:48:12] >>> [1] 12 outlier cells
#> ℹ [2026-02-11 03:48:12] >>> [1] 25low-UMI cells
#> ℹ [2026-02-11 03:48:12] >>> [1] 251low-gene cells
#> ℹ [2026-02-11 03:48:12] >>> [1] 0high-mito cells
#> ℹ [2026-02-11 03:48:12] >>> [1] 0high-ribo cells
#> ℹ [2026-02-11 03:48:12] >>> [1] 0ribo_mito_ratio outlier cells
#> ℹ [2026-02-11 03:48:12] >>> [1] 0species-contaminated cells
#> ℹ [2026-02-11 03:48:12] >>> Remained cells after filtering: [1] 692
CellStatPlot(
srt = ifnb_sub,
stat.by = c(
"db_qc", "outlier_qc",
"umi_qc", "gene_qc",
"mito_qc", "ribo_qc",
"ribo_mito_ratio_qc", "species_qc"
),
plot_type = "upset",
stat_level = "Fail"
)
#> ! [2026-02-11 03:48:12] `stat_type` is forcibly set to "count" when plot "sankey", "chord", "venn", and "upset"
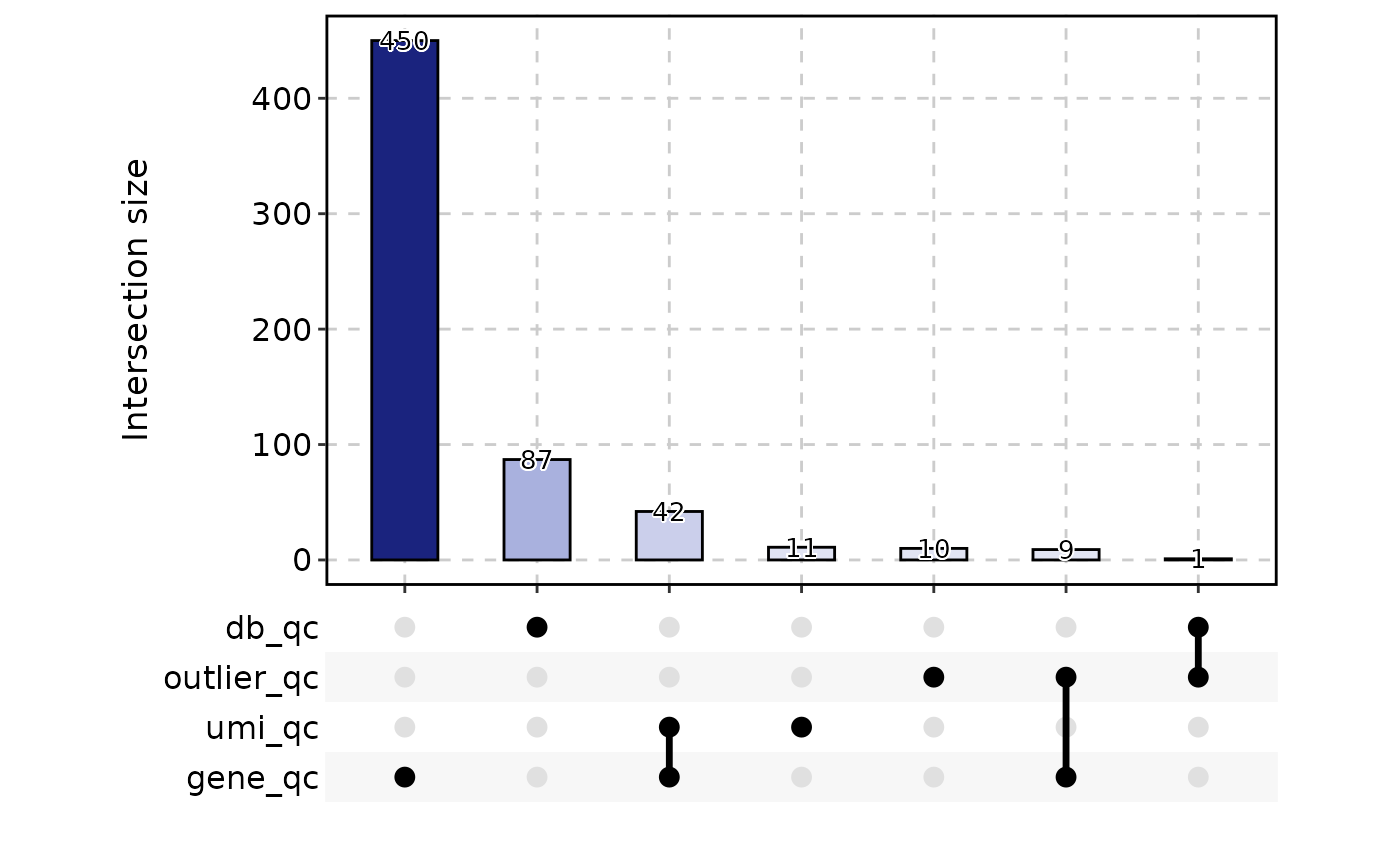 table(ifnb_sub$CellQC)
#>
#> Pass Fail
#> 1382 618
table(ifnb_sub$CellQC)
#>
#> Pass Fail
#> 1382 618