RunCellChat performs CellChat analysis on a Seurat object to investigate cell-to-cell communication. The results are stored in the Seurat object and can be visualized using CellChatPlot.
Usage
RunCellChat(
srt,
group.by,
species = c("Homo_sapiens", "Mus_musculus", "zebrafish"),
split.by = NULL,
annotation_selected = NULL,
group_column = NULL,
group_cmp = NULL,
thresh = 0.05,
min.cells = 10,
verbose = TRUE
)Arguments
- srt
A Seurat object.
- group.by
Name of one or more meta.data columns to group (color) cells by.
- species
The species of the data, either 'human', 'mouse' or 'zebrafish'.
- split.by
Name of a column in meta.data column to split plot by. Default is
NULL.- annotation_selected
A vector of cell annotations of interest for running the CellChat analysis. If not provided, all cell types will be considered.
- group_column
Name of the metadata column in the Seurat object that defines conditions or groups.
- group_cmp
A list of pairwise condition comparisons for differential CellChat analysis.
- thresh
The threshold for computing centrality scores.
- min.cells
the minmum number of expressed cells required for the genes that are considered for cell-cell communication analysis.
- verbose
Whether to print the message. Default is
TRUE.
Examples
data(pancreas_sub)
pancreas_sub <- standard_scop(pancreas_sub)
#> ℹ [2026-02-11 03:40:22] Start standard scop workflow...
#> ℹ [2026-02-11 03:40:22] Checking a list of <Seurat>...
#> ! [2026-02-11 03:40:22] Data 1/1 of the `srt_list` is "unknown"
#> ℹ [2026-02-11 03:40:22] Perform `NormalizeData()` with `normalization.method = 'LogNormalize'` on the data 1/1 of the `srt_list`...
#> ℹ [2026-02-11 03:40:24] Perform `Seurat::FindVariableFeatures()` on the data 1/1 of the `srt_list`...
#> ℹ [2026-02-11 03:40:25] Use the separate HVF from srt_list
#> ℹ [2026-02-11 03:40:25] Number of available HVF: 2000
#> ℹ [2026-02-11 03:40:25] Finished check
#> ℹ [2026-02-11 03:40:25] Perform `Seurat::ScaleData()`
#> ℹ [2026-02-11 03:40:26] Perform pca linear dimension reduction
#> ℹ [2026-02-11 03:40:26] Perform `Seurat::FindClusters()` with `cluster_algorithm = 'louvain'` and `cluster_resolution = 0.6`
#> ℹ [2026-02-11 03:40:27] Reorder clusters...
#> ℹ [2026-02-11 03:40:27] Perform umap nonlinear dimension reduction
#> ℹ [2026-02-11 03:40:27] Non-linear dimensionality reduction (umap) using (Standardpca) dims (1-50) as input
#> ℹ [2026-02-11 03:40:30] Non-linear dimensionality reduction (umap) using (Standardpca) dims (1-50) as input
#> ✔ [2026-02-11 03:40:34] Run scop standard workflow completed
pancreas_sub <- RunCellChat(
pancreas_sub,
group.by = "CellType",
species = "Mus_musculus"
)
#> ℹ [2026-02-11 03:40:34] Start CellChat analysis
#> Registered S3 method overwritten by 'ggnetwork':
#> method from
#> fortify.igraph ggtree
#> [1] "Create a CellChat object from a data matrix"
#> Set cell identities for the new CellChat object
#> The cell groups used for CellChat analysis are Ductal, Ngn3-high-EP, Endocrine, Ngn3-low-EP, Pre-endocrine
#> The number of highly variable ligand-receptor pairs used for signaling inference is 841
#> triMean is used for calculating the average gene expression per cell group.
#> [1] ">>> Run CellChat on sc/snRNA-seq data <<< [2026-02-11 03:41:48.396036]"
#> [1] ">>> CellChat inference is done. Parameter values are stored in `object@options$parameter` <<< [2026-02-11 03:42:58.202989]"
#> ✔ [2026-02-11 03:42:58] CellChat analysis completed
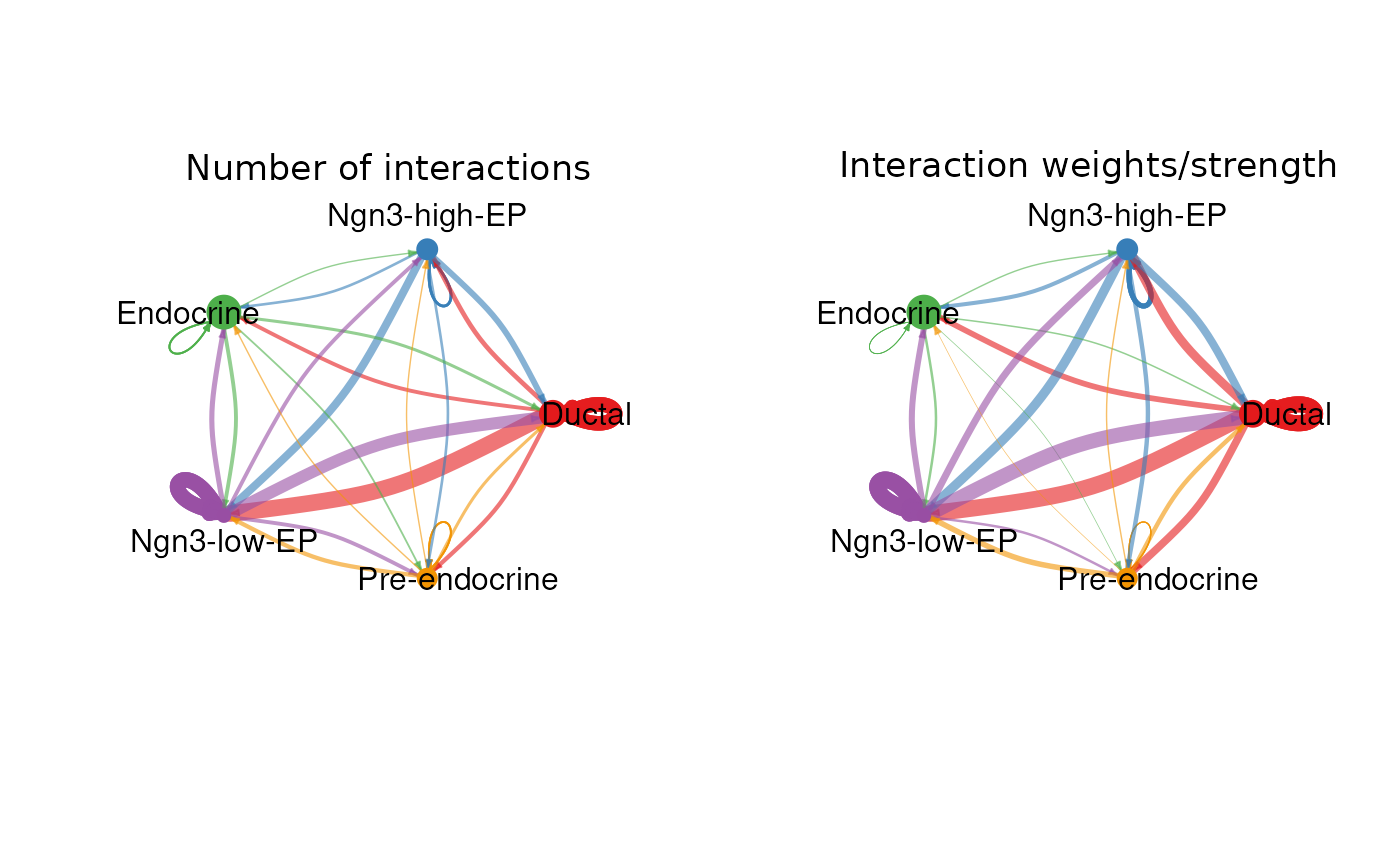
CellChatPlot(pancreas_sub)
#> ℹ [2026-02-11 03:42:58] Creating "aggregate" plot for condition "ALL"
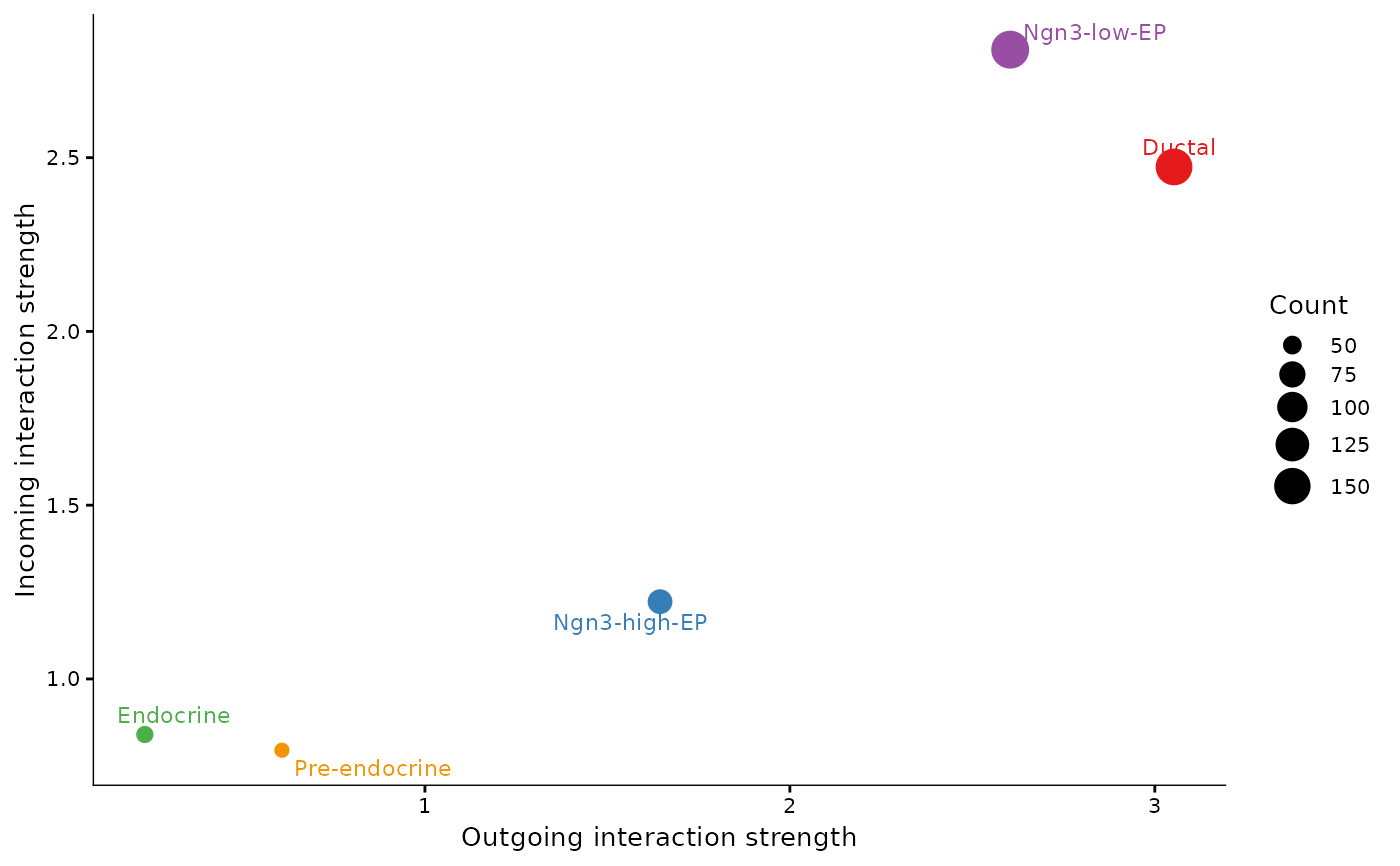
 #> Signaling role analysis on the aggregated cell-cell communication network from all signaling pathways
#> Signaling role analysis on the aggregated cell-cell communication network from all signaling pathways
 #> ✔ [2026-02-11 03:42:58] Plot creation completed
#> ✔ [2026-02-11 03:42:58] Plot creation completed