This function subsamples a matrix using either random sampling or meta cells method.
Usage
subsampling(
matrix,
subsampling_method = c("sample", "meta_cells", "pseudobulk"),
subsampling_ratio = 1,
seed = 1,
verbose = TRUE,
...
)Arguments
- matrix
The input matrix to be subsampled.
- subsampling_method
The method to use for subsampling. Options are
"sample","pseudobulk"or"meta_cells".- subsampling_ratio
The percent of all samples used for fit_srm. Default is
1.- seed
The random seed for cross-validation. Default is
1.- verbose
Whether to print progress messages. Default is
TRUE.- ...
Parameters for other methods.
Examples
data(example_matrix)
data("example_ground_truth")
subsample_matrix <- subsampling(
example_matrix,
subsampling_ratio = 0.5
)
#> ℹ [2026-01-23 02:16:18] Subsample matrix generated, dimensions: 2500 cells by 18 genes
subsample_matrix_2 <- subsampling(
example_matrix,
subsampling_method = "meta_cells",
subsampling_ratio = 0.5,
fast_pca = FALSE
)
#> ! [2026-01-23 02:16:18] Number of PCs of PCA result is less than the desired number, using all PCs.
#> ℹ [2026-01-23 02:16:19] Subsample matrix generated, dimensions: 2500 cells by 18 genes
subsample_matrix_3 <- subsampling(
example_matrix,
subsampling_method = "pseudobulk",
subsampling_ratio = 0.5
)
#> ℹ [2026-01-23 02:16:20] Subsample matrix generated, dimensions: 2500 cells by 18 genes
calculate_metrics(
inferCSN(example_matrix),
example_ground_truth,
return_plot = TRUE
)
#> ℹ [2026-01-23 02:16:20] Inferring network for <matrix/array>...
#> ◌ [2026-01-23 02:16:20] Checking parameters...
#> ℹ [2026-01-23 02:16:20] Using L0 sparse regression model
#> ℹ [2026-01-23 02:16:20] Using 1 core
#> ⠙ [2026-01-23 02:16:20] Running for g1 [1/18] ■■■ …
#> ✔ [2026-01-23 02:16:20] Completed 18 tasks in 182ms
#>
#> ℹ [2026-01-23 02:16:20] Building results
#> ✔ [2026-01-23 02:16:20] Inferring network done
#> ℹ [2026-01-23 02:16:20] Network information:
#> ℹ Edges Regulators Targets
#> ℹ 1 306 18 18
#> $metrics
#> Metric Value
#> 1 AUROC 0.952
#> 2 AUPRC 0.437
#> 3 Precision 0.529
#> 4 Recall 1.000
#> 5 F1 0.692
#> 6 ACC 0.948
#> 7 JI 0.514
#> 8 SI 18.000
#>
#> $plot
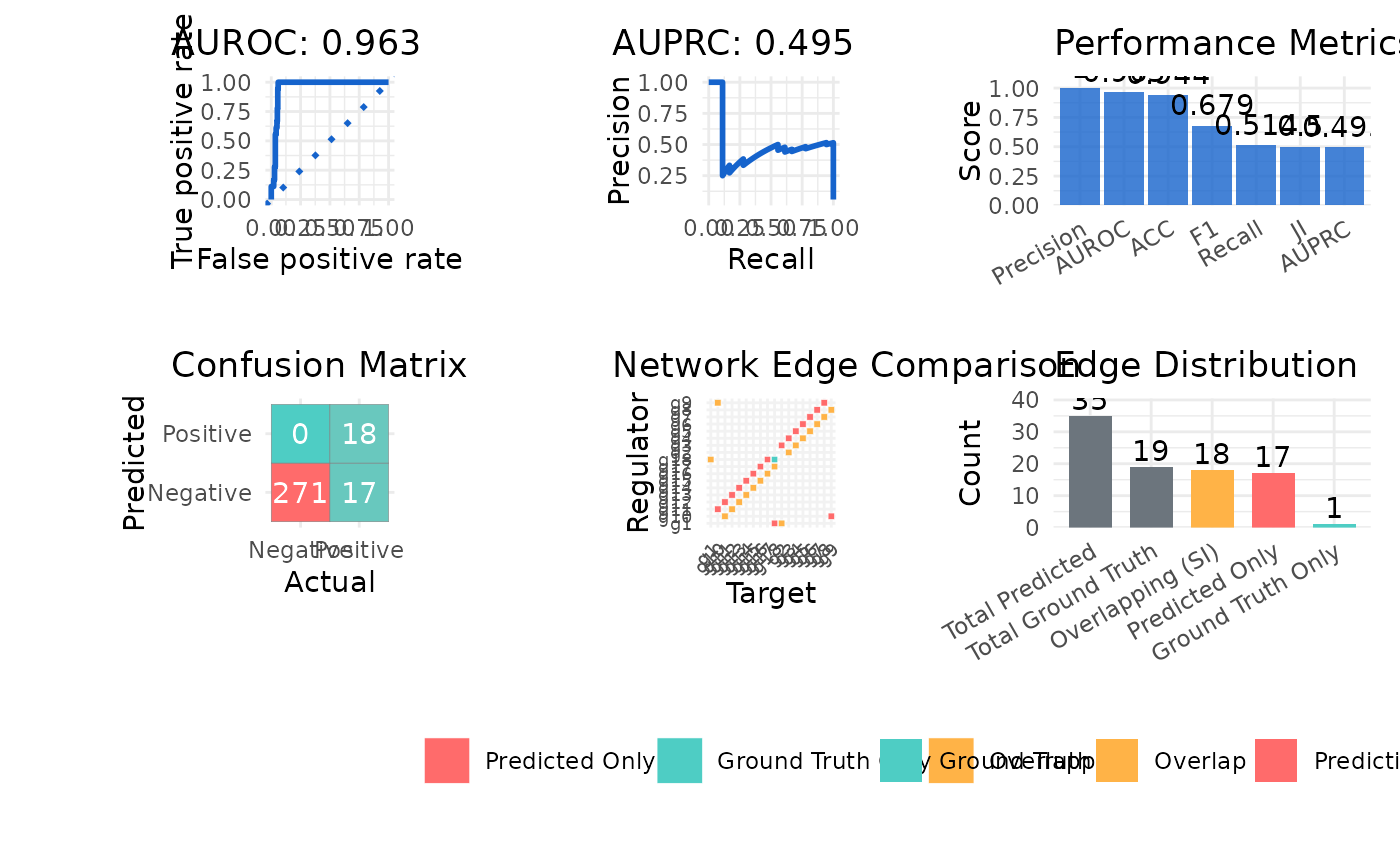 #>
calculate_metrics(
inferCSN(subsample_matrix),
example_ground_truth,
return_plot = TRUE
)
#> ℹ [2026-01-23 02:16:20] Inferring network for <matrix/array>...
#> ◌ [2026-01-23 02:16:20] Checking parameters...
#> ℹ [2026-01-23 02:16:20] Using L0 sparse regression model
#> ℹ [2026-01-23 02:16:20] Using 1 core
#> ℹ [2026-01-23 02:16:20] Building results
#> ✔ [2026-01-23 02:16:20] Inferring network done
#> ℹ [2026-01-23 02:16:20] Network information:
#> ℹ Edges Regulators Targets
#> ℹ 1 306 18 18
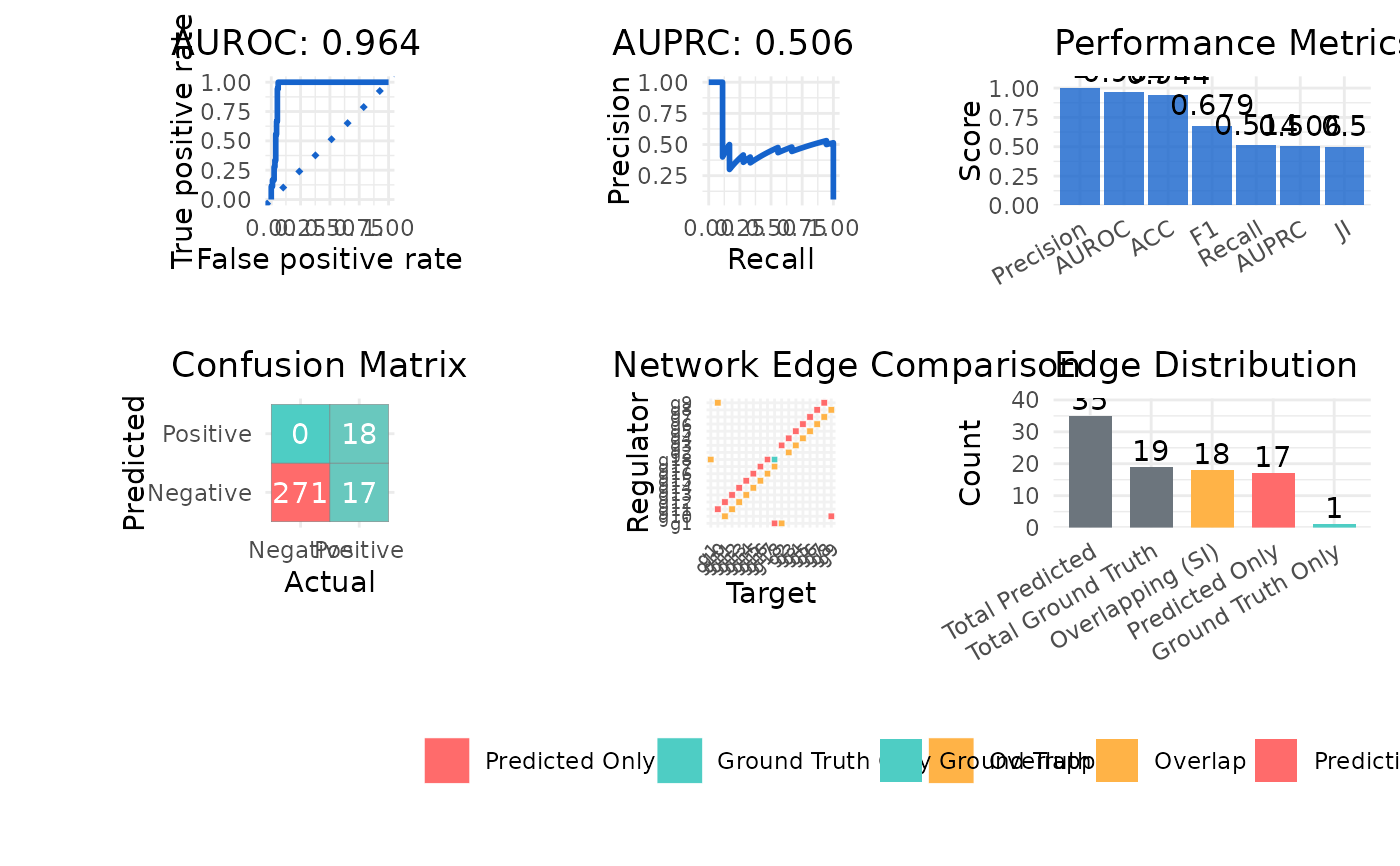
#> $metrics
#> Metric Value
#> 1 AUROC 0.955
#> 2 AUPRC 0.449
#> 3 Precision 0.529
#> 4 Recall 1.000
#> 5 F1 0.692
#> 6 ACC 0.948
#> 7 JI 0.514
#> 8 SI 18.000
#>
#> $plot
#>
calculate_metrics(
inferCSN(subsample_matrix),
example_ground_truth,
return_plot = TRUE
)
#> ℹ [2026-01-23 02:16:20] Inferring network for <matrix/array>...
#> ◌ [2026-01-23 02:16:20] Checking parameters...
#> ℹ [2026-01-23 02:16:20] Using L0 sparse regression model
#> ℹ [2026-01-23 02:16:20] Using 1 core
#> ℹ [2026-01-23 02:16:20] Building results
#> ✔ [2026-01-23 02:16:20] Inferring network done
#> ℹ [2026-01-23 02:16:20] Network information:
#> ℹ Edges Regulators Targets
#> ℹ 1 306 18 18
#> $metrics
#> Metric Value
#> 1 AUROC 0.955
#> 2 AUPRC 0.449
#> 3 Precision 0.529
#> 4 Recall 1.000
#> 5 F1 0.692
#> 6 ACC 0.948
#> 7 JI 0.514
#> 8 SI 18.000
#>
#> $plot
 #>
calculate_metrics(
inferCSN(subsample_matrix_2),
example_ground_truth,
return_plot = TRUE
)
#> ℹ [2026-01-23 02:16:20] Inferring network for <matrix/array>...
#> ◌ [2026-01-23 02:16:20] Checking parameters...
#> ℹ [2026-01-23 02:16:20] Using L0 sparse regression model
#> ℹ [2026-01-23 02:16:20] Using 1 core
#> ℹ [2026-01-23 02:16:20] Building results
#> ✔ [2026-01-23 02:16:20] Inferring network done
#> ℹ [2026-01-23 02:16:20] Network information:
#> ℹ Edges Regulators Targets
#> ℹ 1 306 18 18
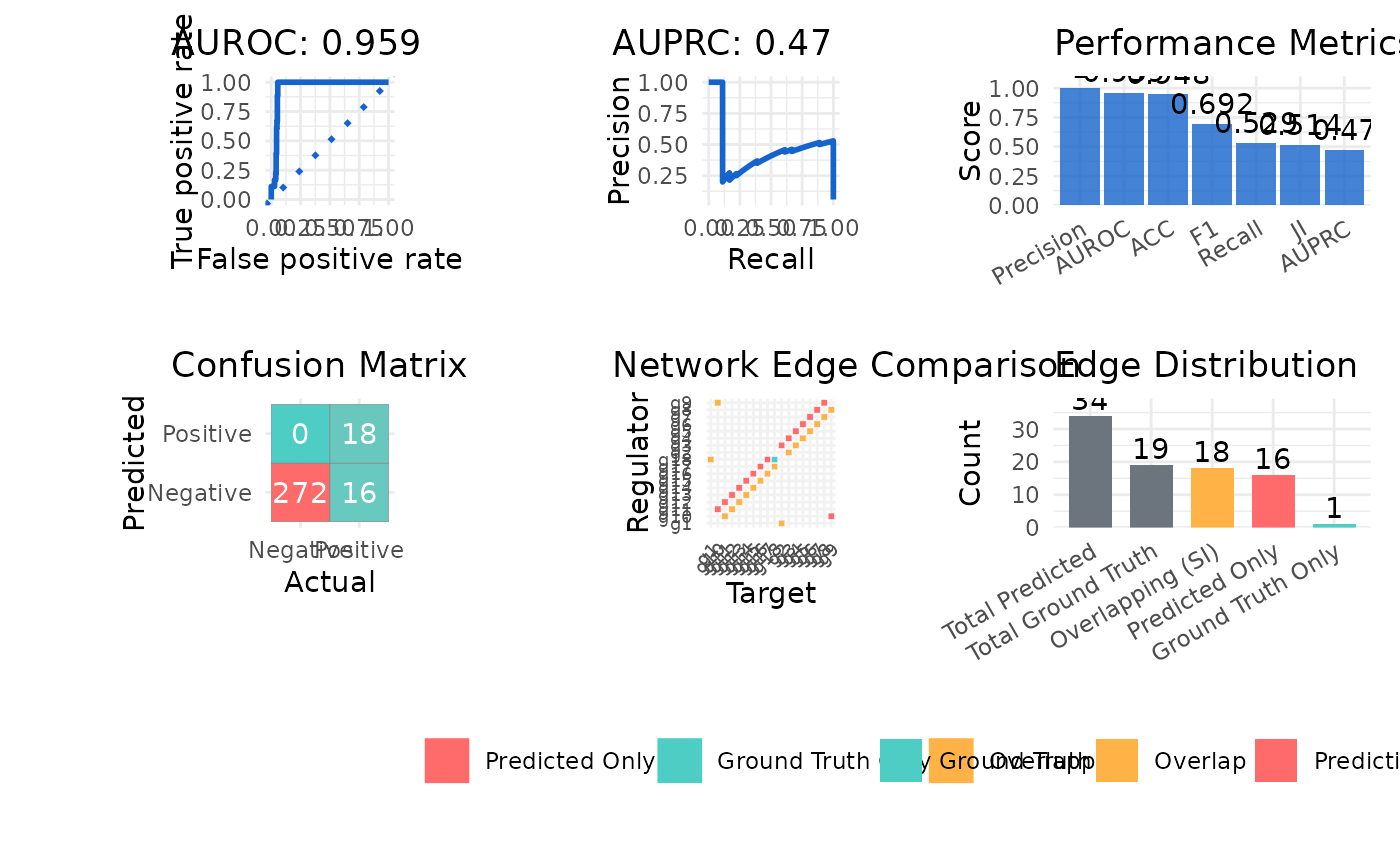
#> $metrics
#> Metric Value
#> 1 AUROC 0.952
#> 2 AUPRC 0.439
#> 3 Precision 0.529
#> 4 Recall 1.000
#> 5 F1 0.692
#> 6 ACC 0.948
#> 7 JI 0.514
#> 8 SI 18.000
#>
#> $plot
#>
calculate_metrics(
inferCSN(subsample_matrix_2),
example_ground_truth,
return_plot = TRUE
)
#> ℹ [2026-01-23 02:16:20] Inferring network for <matrix/array>...
#> ◌ [2026-01-23 02:16:20] Checking parameters...
#> ℹ [2026-01-23 02:16:20] Using L0 sparse regression model
#> ℹ [2026-01-23 02:16:20] Using 1 core
#> ℹ [2026-01-23 02:16:20] Building results
#> ✔ [2026-01-23 02:16:20] Inferring network done
#> ℹ [2026-01-23 02:16:20] Network information:
#> ℹ Edges Regulators Targets
#> ℹ 1 306 18 18
#> $metrics
#> Metric Value
#> 1 AUROC 0.952
#> 2 AUPRC 0.439
#> 3 Precision 0.529
#> 4 Recall 1.000
#> 5 F1 0.692
#> 6 ACC 0.948
#> 7 JI 0.514
#> 8 SI 18.000
#>
#> $plot
 #>
calculate_metrics(
inferCSN(subsample_matrix_3),
example_ground_truth,
return_plot = TRUE
)
#> ℹ [2026-01-23 02:16:21] Inferring network for <matrix/array>...
#> ◌ [2026-01-23 02:16:21] Checking parameters...
#> ℹ [2026-01-23 02:16:21] Using L0 sparse regression model
#> ℹ [2026-01-23 02:16:21] Using 1 core
#> ℹ [2026-01-23 02:16:21] Building results
#> ✔ [2026-01-23 02:16:21] Inferring network done
#> ℹ [2026-01-23 02:16:21] Network information:
#> ℹ Edges Regulators Targets
#> ℹ 1 306 18 18
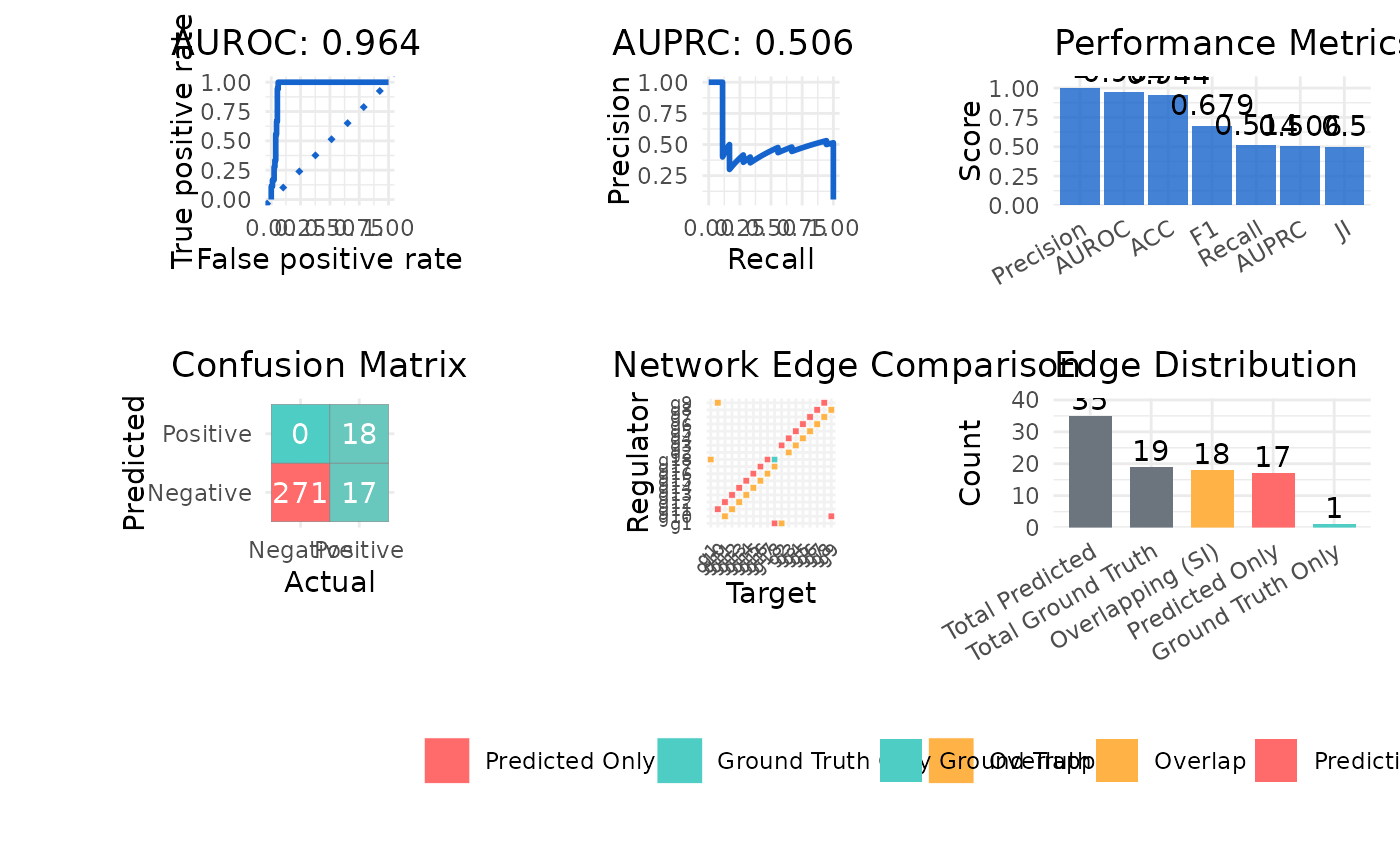
#> $metrics
#> Metric Value
#> 1 AUROC 0.955
#> 2 AUPRC 0.449
#> 3 Precision 0.529
#> 4 Recall 1.000
#> 5 F1 0.692
#> 6 ACC 0.948
#> 7 JI 0.514
#> 8 SI 18.000
#>
#> $plot
#>
calculate_metrics(
inferCSN(subsample_matrix_3),
example_ground_truth,
return_plot = TRUE
)
#> ℹ [2026-01-23 02:16:21] Inferring network for <matrix/array>...
#> ◌ [2026-01-23 02:16:21] Checking parameters...
#> ℹ [2026-01-23 02:16:21] Using L0 sparse regression model
#> ℹ [2026-01-23 02:16:21] Using 1 core
#> ℹ [2026-01-23 02:16:21] Building results
#> ✔ [2026-01-23 02:16:21] Inferring network done
#> ℹ [2026-01-23 02:16:21] Network information:
#> ℹ Edges Regulators Targets
#> ℹ 1 306 18 18
#> $metrics
#> Metric Value
#> 1 AUROC 0.955
#> 2 AUPRC 0.449
#> 3 Precision 0.529
#> 4 Recall 1.000
#> 5 F1 0.692
#> 6 ACC 0.948
#> 7 JI 0.514
#> 8 SI 18.000
#>
#> $plot
 #>
#>