Plot dynamic networks
Usage
plot_dynamic_networks(
network_table,
celltypes_order,
ntop = 10,
title = NULL,
theme_type = "theme_void",
plot_type = "ggplot",
layout = "fruchtermanreingold",
nrow = 2,
figure_save = FALSE,
figure_name = NULL,
figure_width = 6,
figure_height = 6,
seed = 1
)Arguments
- network_table
The weight data table of network.
- celltypes_order
The order of cell types.
- ntop
The number of top genes to plot. Default is
10.- title
The title of figure. Default is
NULL.- theme_type
The theme of figure. Could be
"theme_void","theme_blank", or"theme_facet". Default is"theme_void".- plot_type
The type of figure. Could be
"ggplot","animate", or"ggplotly". Default is"ggplot".- layout
The layout of figure. Could be
"fruchtermanreingold"or"kamadakawai". Default is"fruchtermanreingold".- nrow
The number of rows of figure. Default is
2.- figure_save
Whether to save the figure file. Default is
FALSE.- figure_name
The name of figure file. Default is
NULL.- figure_width
The width of figure. Default is
6.- figure_height
The height of figure. Default is
6.- seed
The seed random use to plot network. Default is
1.
Examples
data(example_matrix)
network <- inferCSN(example_matrix)[1:100, ]
#> ℹ [2026-01-23 02:16:02] Inferring network for <matrix/array>...
#> ◌ [2026-01-23 02:16:02] Checking parameters...
#> ℹ [2026-01-23 02:16:02] Using L0 sparse regression model
#> ℹ [2026-01-23 02:16:02] Using 1 core
#> ⠙ [2026-01-23 02:16:02] Running for g1 [1/18] ■■■ …
#> ✔ [2026-01-23 02:16:02] Completed 18 tasks in 169ms
#>
#> ℹ [2026-01-23 02:16:02] Building results
#> ✔ [2026-01-23 02:16:02] Inferring network done
#> ℹ [2026-01-23 02:16:02] Network information:
#> ℹ Edges Regulators Targets
#> ℹ 1 306 18 18
network$celltype <- c(
rep("cluster1", 20),
rep("cluster2", 20),
rep("cluster3", 20),
rep("cluster5", 20),
rep("cluster6", 20)
)
celltypes_order <- c(
"cluster5", "cluster3",
"cluster2", "cluster1",
"cluster6"
)
plot_dynamic_networks(
network,
celltypes_order = celltypes_order
)
#> Warning: Using `size` aesthetic for lines was deprecated in ggplot2 3.4.0.
#> ℹ Please use `linewidth` instead.
#> ℹ The deprecated feature was likely used in the inferCSN package.
#> Please report the issue at <https://github.com/mengxu98/inferCSN/issues>.
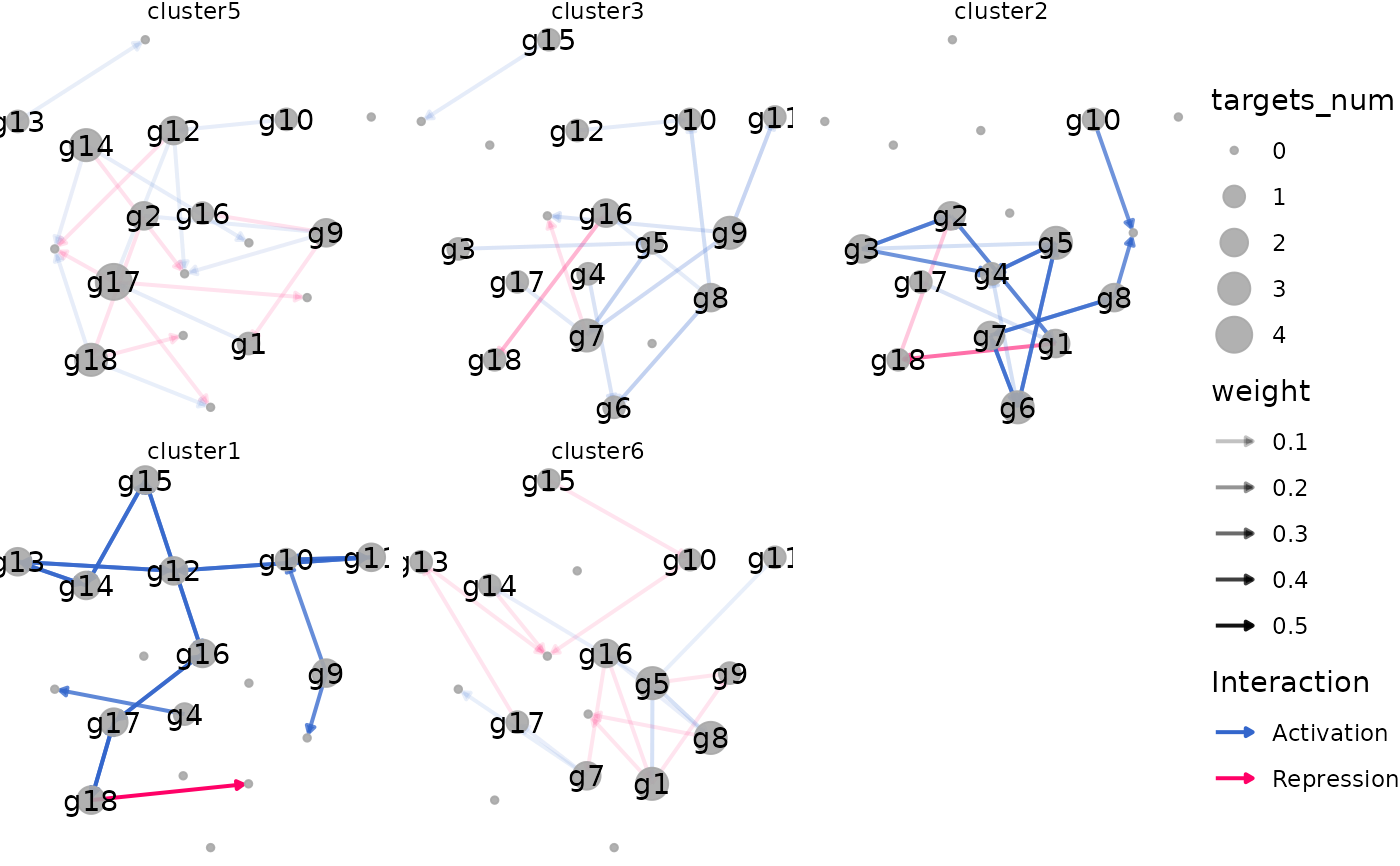 plot_dynamic_networks(
network,
celltypes_order = celltypes_order[1:3]
)
plot_dynamic_networks(
network,
celltypes_order = celltypes_order[1:3]
)
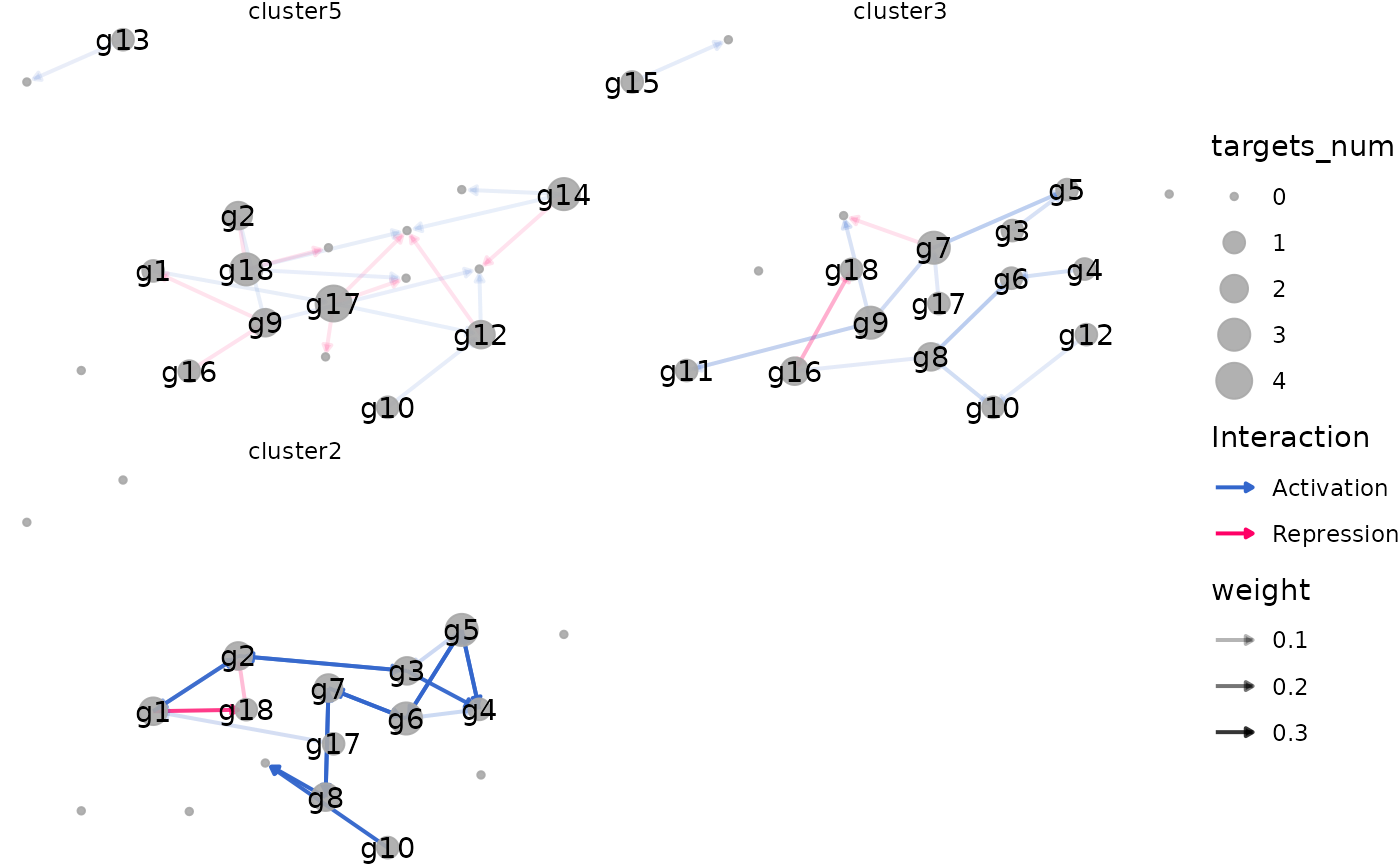 plot_dynamic_networks(
network,
celltypes_order = celltypes_order,
plot_type = "ggplotly"
)
plot_dynamic_networks(
network,
celltypes_order = celltypes_order,
plot_type = "ggplotly"
)